Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence - PubMed (original) (raw)
Review
Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence
Eugene V Koonin. Biol Direct. 2017.
Abstract
Complementarity between nucleic acid molecules is central to biological information transfer processes. Apart from the basal processes of replication, transcription and translation, complementarity is also employed by multiple defense and regulatory systems. All cellular life forms possess defense systems against viruses and mobile genetic elements, and in most of them some of the defense mechanisms involve small guide RNAs or DNAs that recognize parasite genomes and trigger their inactivation. The nucleic acid-guided defense systems include prokaryotic Argonaute (pAgo)-centered innate immunity and CRISPR-Cas adaptive immunity as well as diverse branches of RNA interference (RNAi) in eukaryotes. The archaeal pAgo machinery is the direct ancestor of eukaryotic RNAi that, however, acquired additional components, such as Dicer, and enormously diversified through multiple duplications. In contrast, eukaryotes lack any heritage of the CRISPR-Cas systems, conceivably, due to the cellular toxicity of some Cas proteins that would get activated as a result of operon disruption in eukaryotes. The adaptive immunity function in eukaryotes is taken over partly by the PIWI RNA branch of RNAi and partly by protein-based immunity. In this review, I briefly discuss the interplay between homology and analogy in the evolution of RNA- and DNA-guided immunity, and attempt to formulate some general evolutionary principles for this ancient class of defense systems.
Reviewers: This article was reviewed by Mikhail Gelfand and Bojan Zagrovic.
Figures
Fig. 1
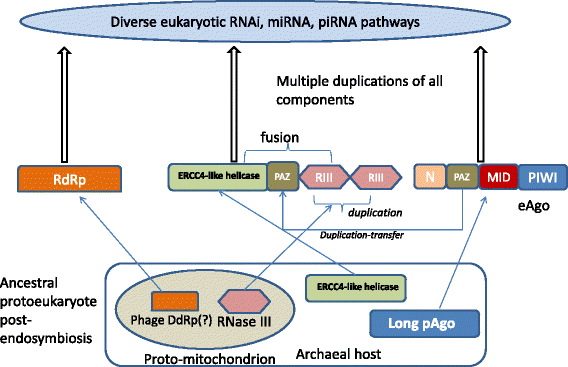
The evolutionary history of eukaryotic RNAi: assembly from diverse archaeal and bacterial ancestors. The “bacterial” and “archaeal” components of the RNAi protein machinery are assumed to have evolved from the proto-mitochondrial endosymbiont and its archaeal host, respectively. This scenario rests on the fact that RNase III is a protein that is nearly ubiquitous in bacteria but rare in archaea, and the (DNA-dependent) RNA polymerase that is thought to be the ancestor of the RNAi RdRp so far has been identified only in bacteriophages (not in archaeal viruses). However, it cannot be ruled out that these genes have been acquired by the mesophilic archaeal ancestor of eukaryotes (presumably, a member of the Lokiarchaeota) prior to endosymbiosis. RIII, RNAse III
Fig. 2
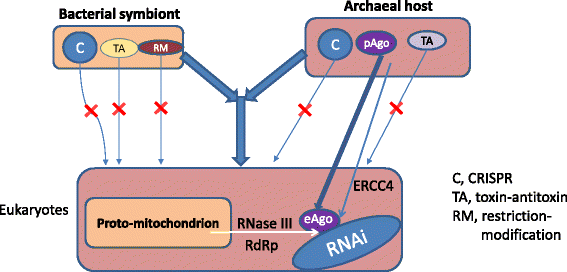
The fates of prokaryotic defense systems in eukaryotes. C, CRISPR-Cas; RM, restriction-modification; TA, toxins-antitoxins
Fig. 3
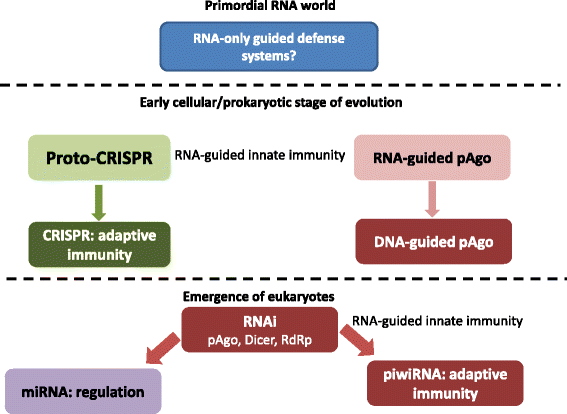
Evolution of RNA/DNA-guided defense and regulatory systems: from the RNA world to the present
Similar articles
- Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements.
Makarova KS, Wolf YI, van der Oost J, Koonin EV. Makarova KS, et al. Biol Direct. 2009 Aug 25;4:29. doi: 10.1186/1745-6150-4-29. Biol Direct. 2009. PMID: 19706170 Free PMC article. - DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins.
Lisitskaya L, Aravin AA, Kulbachinskiy A. Lisitskaya L, et al. Nat Commun. 2018 Dec 4;9(1):5165. doi: 10.1038/s41467-018-07449-7. Nat Commun. 2018. PMID: 30514832 Free PMC article. Review. - [The CRISPR case, « ready-made » mutations and Lamarckian evolution of an adaptive immunity system].
Casane D, Laurenti P. Casane D, et al. Med Sci (Paris). 2016 Jun-Jul;32(6-7):640-5. doi: 10.1051/medsci/20163206029. Epub 2016 Jul 12. Med Sci (Paris). 2016. PMID: 27406776 French. - Diverse CRISPR-Cas Complexes Require Independent Translation of Small and Large Subunits from a Single Gene.
McBride TM, Schwartz EA, Kumar A, Taylor DW, Fineran PC, Fagerlund RD. McBride TM, et al. Mol Cell. 2020 Dec 17;80(6):971-979.e7. doi: 10.1016/j.molcel.2020.11.003. Epub 2020 Nov 27. Mol Cell. 2020. PMID: 33248026 - Evolutionary Genomics of Defense Systems in Archaea and Bacteria.
Koonin EV, Makarova KS, Wolf YI. Koonin EV, et al. Annu Rev Microbiol. 2017 Sep 8;71:233-261. doi: 10.1146/annurev-micro-090816-093830. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657885 Free PMC article. Review.
Cited by
- Bacterial Argonaute nucleases reveal different modes of DNA targeting in vitro and in vivo.
Lisitskaya L, Kropocheva E, Agapov A, Prostova M, Panteleev V, Yudin D, Ryazansky S, Kuzmenko A, Aravin AA, Esyunina D, Kulbachinskiy A. Lisitskaya L, et al. Nucleic Acids Res. 2023 Jun 9;51(10):5106-5124. doi: 10.1093/nar/gkad290. Nucleic Acids Res. 2023. PMID: 37094066 Free PMC article. - Inevitability of the emergence and persistence of genetic parasites caused by evolutionary instability of parasite-free states.
Koonin EV, Wolf YI, Katsnelson MI. Koonin EV, et al. Biol Direct. 2017 Dec 4;12(1):31. doi: 10.1186/s13062-017-0202-5. Biol Direct. 2017. PMID: 29202832 Free PMC article. - Non-Coding RNA in Tumor Cells and Tumor-Associated Myeloid Cells-Function and Therapeutic Potential.
Binder AK, Bremm F, Dörrie J, Schaft N. Binder AK, et al. Int J Mol Sci. 2024 Jul 2;25(13):7275. doi: 10.3390/ijms25137275. Int J Mol Sci. 2024. PMID: 39000381 Free PMC article. Review. - Viperin-like proteins interfere with RNA viruses in plants.
Kamel R, Aman R, Mahfouz MM. Kamel R, et al. Front Plant Sci. 2024 Jun 4;15:1385169. doi: 10.3389/fpls.2024.1385169. eCollection 2024. Front Plant Sci. 2024. PMID: 38895613 Free PMC article.
References
- Schroedinger E. What Is Life? with “Mind and Matter” and “Autobiographical Sketches”. Cambridge: Cambridge University Press; 2012.
- Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. Upper Saddle River, NJ: FT press; 2011.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources