NOVOPlasty: de novo assembly of organelle genomes from whole genome data - PubMed (original) (raw)
NOVOPlasty: de novo assembly of organelle genomes from whole genome data
Nicolas Dierckxsens et al. Nucleic Acids Res. 2017.
Abstract
The evolution in next-generation sequencing (NGS) technology has led to the development of many different assembly algorithms, but few of them focus on assembling the organelle genomes. These genomes are used in phylogenetic studies, food identification and are the most deposited eukaryotic genomes in GenBank. Producing organelle genome assembly from whole genome sequencing (WGS) data would be the most accurate and least laborious approach, but a tool specifically designed for this task is lacking. We developed a seed-and-extend algorithm that assembles organelle genomes from whole genome sequencing (WGS) data, starting from a related or distant single seed sequence. The algorithm has been tested on several new (Gonioctena intermedia and Avicennia marina) and public (Arabidopsis thaliana and Oryza sativa) whole genome Illumina data sets where it outperforms known assemblers in assembly accuracy and coverage. In our benchmark, NOVOPlasty assembled all tested circular genomes in less than 30 min with a maximum memory requirement of 16 GB and an accuracy over 99.99%. In conclusion, NOVOPlasty is the sole de novo assembler that provides a fast and straightforward extraction of the extranuclear genomes from WGS data in one circular high quality contig. The software is open source and can be downloaded at https://github.com/ndierckx/NOVOPlasty.
Figures
Figure 1.
Coverage depth for a 12 000 bp long region of the mitochondrial genome of Gonioctena intermedia. There are several regions with a low GC content, resulting in a reduced read coverage.
Figure 2.
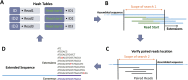
Work flow of NOVOPlasty. For simplicity the work flow was limited to unidirectional extension. (A) All reads are stored in a hash table with a unique id. A second hash table contains the ids for the read start = k-mer parameter (default = 38) of the corresponding read. (B) Scope of search 1 is the region where a match of the ‘read start’ indicates a extension of the sequence. All these matching reads are stored separately. (C) The position of the paired reads are verified by aligning each paired read to a previous assembled area, which is determined by the library insert size (scope of search 2). (D) A consensus sequence of the different extensions is determined.
Figure 3.
Comparison between the NOVOPlasty and CLC alignments of three different chloroplast assemblies against their respective reference. (A) CLC and NOVOPlasty assemblies of SRR1174256 (A. thaliana) against GenBank entry AP000423.1. (B) CLC and NOVOPlasty assemblies of ERR477442 (O. sativa) against GenBank entry KM088022.1. (C) CLC assembly of A. marina against the manually inspected NOVOPlasty assembly.
Figure 4.
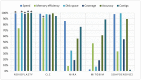
Score graph derived from the benchmark study. Each property of each assembler was given a score proportional to the other assemblers. Each score was based on the average results of seven assemblies and expressed in percentage. A score of 100% is always seen as most favorable, more detailed explanation can be found in the ‘Quality assessment’ section of Materials and Methods. (*) Highest score for the corresponding property.
Figure 5.
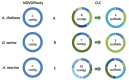
Seed compatibility test for the de novo assembly of the human mitochondrium with 12 different mitochondrial genomes as seed sequence. A green dot means that the mitochondrial genome of that species can be used as a seed for the mitochondrial assembly of H. sapiens. Red X means unsuccessful. Phylogenetic tree based on information extracted from the NCBI taxonomy database (20), using phyloT (
).
Figure 6.
Seed compatibility test for the de novo assembly of the chloroplast from Arabidopsis thaliana with 12 different chloroplast genomes and 12 subunits (RuBP) as a seed sequence. A green dot means that the chloroplast genome of that species can be used as a seed for the chloroplast assembly of A. thaliana. Red M indicates that NOVOPlasty assembled the mitochondrial genome instead of the chloroplast genome. Same color indications for the RuBP unit. Phylogenetic tree based on information extracted from the NCBI taxonomy database (20), using phyloT (
).
Similar articles
- Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data.
Soorni A, Haak D, Zaitlin D, Bombarely A. Soorni A, et al. BMC Genomics. 2017 Jan 7;18(1):49. doi: 10.1186/s12864-016-3412-9. BMC Genomics. 2017. PMID: 28061749 Free PMC article. - Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data.
Al-Nakeeb K, Petersen TN, Sicheritz-Pontén T. Al-Nakeeb K, et al. BMC Bioinformatics. 2017 Nov 21;18(1):510. doi: 10.1186/s12859-017-1927-y. BMC Bioinformatics. 2017. PMID: 29162031 Free PMC article. - GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes.
Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. Jin JJ, et al. Genome Biol. 2020 Sep 10;21(1):241. doi: 10.1186/s13059-020-02154-5. Genome Biol. 2020. PMID: 32912315 Free PMC article. - The present and future of de novo whole-genome assembly.
Sohn JI, Nam JW. Sohn JI, et al. Brief Bioinform. 2018 Jan 1;19(1):23-40. doi: 10.1093/bib/bbw096. Brief Bioinform. 2018. PMID: 27742661 Review. - [Mitogenome assembly strategies and software applications in the genome era].
Kuang WM, Yu L. Kuang WM, et al. Yi Chuan. 2019 Nov 20;41(11):979-993. doi: 10.16288/j.yczz.19-227. Yi Chuan. 2019. PMID: 31735702 Review. Chinese.
Cited by
- The Complete Mitochondrial Genome of Nephropsis grandis: Insights into the Phylogeny of Nephropidae Mitochondrial Genome.
Liang X, Sun Y, Chen J, Li J, Ye Y. Liang X, et al. Biochem Genet. 2024 Oct 29. doi: 10.1007/s10528-024-10948-6. Online ahead of print. Biochem Genet. 2024. PMID: 39470934 - The complete mitochondrial genome of the shining leaf chafer Mimela junii (Duftschmidt, 1805) (Coleoptera: Scarabaeidae).
Nardi F, Funari R, Carapelli A, Badano D, Frati F, Cucini C. Nardi F, et al. Mitochondrial DNA B Resour. 2024 Oct 23;9(10):1439-1444. doi: 10.1080/23802359.2024.2417936. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 39450202 Free PMC article. - The complete chloroplast genome sequence of Ilex chinensis Sims. (Aquifoliaceae), a folk herbal medicine plant in China.
Zhou Y, Li N, Chen H, Chong X, Li Y, Lu X, Zhou T, Zhang F. Zhou Y, et al. Mitochondrial DNA B Resour. 2021 Mar 26;6(3):1241-1242. doi: 10.1080/23802359.2021.1904800. Mitochondrial DNA B Resour. 2021. PMID: 33829094 Free PMC article. - Comparative Analysis of Complete Chloroplast Genome Sequences in Edgeworthia (Thymelaeaceae) and New Insights Into Phylogenetic Relationships.
Qian S, Zhang Y, Lee SY. Qian S, et al. Front Genet. 2021 Mar 10;12:643552. doi: 10.3389/fgene.2021.643552. eCollection 2021. Front Genet. 2021. PMID: 33790948 Free PMC article. - Deciphering the Plastomic Code of Chinese Hog-Peanut (Amphicarpaea edgeworthii Benth., Leguminosae): Comparative Genomics and Evolutionary Insights within the Phaseoleae Tribe.
Xiang YN, Wang XQ, Ding LL, Bai XY, Feng YQ, Qi ZC, Sun YT, Yan XL. Xiang YN, et al. Genes (Basel). 2024 Jan 11;15(1):88. doi: 10.3390/genes15010088. Genes (Basel). 2024. PMID: 38254977 Free PMC article.
References
- Bignell G.R., Miller A.R., Evans I.H.. Isolation of mitochondrial DNA. Methods Mol. Biol. 1996; 53:109–106. - PubMed
- Jansen R.K., Raubeson L.A., Boore J.L., dePamphilis C.W., Chumley T.W., Haberle R.C., Wyman S.K., Alverson A.J., Peery R., Herman S.J.. Elizabeth AZ, Eric HR. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology. 2005; Academic Press; 348–384. - PubMed
- Khan A., Khan I.A, Asif H., Azim M.K.. Current trends in chloroplast genome research. Afr. J. Biotechnol. 2010; 9:3494–3500.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources