In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni - PubMed (original) (raw)
doi: 10.1038/ncomms14500.
Taeyoung Koo 1 3, Sung Wook Park 4 5, Daesik Kim 1 6, Kyoungmi Kim 1, Hee-Yeon Cho 1, Dong Woo Song 2, Kyu Jun Lee 2, Min Hee Jung 2, Seokjoong Kim 2, Jin Hyoung Kim 4 5, Jeong Hun Kim 4 5 7, Jin-Soo Kim 1 3 6
Affiliations
- PMID: 28220790
- PMCID: PMC5473640
- DOI: 10.1038/ncomms14500
In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni
Eunji Kim et al. Nat Commun. 2017.
Abstract
Several CRISPR-Cas9 orthologues have been used for genome editing. Here, we present the smallest Cas9 orthologue characterized to date, derived from Campylobacter jejuni (CjCas9), for efficient genome editing in vivo. After determining protospacer-adjacent motif (PAM) sequences and optimizing single-guide RNA (sgRNA) length, we package the CjCas9 gene, its sgRNA sequence, and a marker gene in an all-in-one adeno-associated virus (AAV) vector and produce the resulting virus at a high titer. CjCas9 is highly specific, cleaving only a limited number of sites in the human or mouse genome. CjCas9, delivered via AAV, induces targeted mutations at high frequencies in mouse muscle cells or retinal pigment epithelium (RPE) cells. Furthermore, CjCas9 targeted to the Vegfa or Hif1a gene in RPE cells reduces the size of laser-induced choroidal neovascularization, suggesting that in vivo genome editing with CjCas9 is a new option for the treatment of age-related macular degeneration.
Conflict of interest statement
J.-S.K. is a co-founder and shareholder of ToolGen, Inc. D.W.S., K.J.L., M.H.J. and S.K. are employed by ToolGen, Inc. Je.H.K., S.W.P., D.W.S., E.K., and S.K. have filed patent applications based on this work. The remaining authors declare no competing financial interests.
Figures
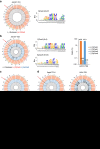
Figure 1. CjCas9 and its PAM specificity.
(a) Cas9 orthologues and their sgRNAs. sgRNA structures were obtained using the mfold web server. See also Supplementary Fig. 1. (b) Schematic showing a PAM assay for characterization of PAM sequences recognized by Cas9. PCR amplicons containing a Cas9 target sequence (yellow) followed by randomized 10-bp (N10) sequences (green) were cleaved by CjCas9 in vitro. The sequence logo showing the PAM specificity was obtained using deep-sequencing data. (c) PAM specificity in cells. Surrogate reporters encoding GFP and RFP and containing a CjCas9 target sequence with degenerate PAMs were transfected with CjCas9 and its sgRNA plasmid into HEK 293 cells. The percentage of cells that express both GFP and RFP was normalized with that obtained using the optimal reporter containing the 5'-NNNNACAC-3' PAM sequence. (−) represents a mock control. Error bars indicate s.e.m. (_n_=3).
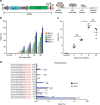
Figure 2. Optimization of sgRNA length for CjCas9.
(a) sgRNAs with variable lengths (19 to 23 nucleotide complementary with a target DNA sequence) were designed and transfected with CjCas9 plasmid into human HEK 293 cells. Genomic DNA was isolated 48 h after transfection. Indel frequencies were analysed by targeted deep sequencing. The first guanine nucleotide at the 5' end that does not match the target sequence is shown in lower case. PAM motifs are shown in red. Error bars indicate s.e.m. (_n_=3). (b) Mutation frequencies at target sites in the mouse genome. Rosa26 and _Tp53_-specific gX22 guide RNAs were designed and transfected into mouse NIH 3T3 cells together with CjCas9 plasmid. Genome editing efficiencies were examined by deep sequencing using genome DNA isolated from cells after 48 h of transfection. PAM motifs are shown in red. Error bars indicate s.e.m. (_n_=3). See also Supplementary Fig. 2. (c) CjCas9-mediated genome editing at the human AAVS1 locus with different PAM sequences. sgRNAs targeting sites with a 5'-NNNNACAC-3' PAM (green; 12 sgRNAs), 5′-NNNNATAC-3′ PAM (light blue; 7 sgRNAs), 5′-NNNNGCAC-3′ PAM (dark blue; 10 sgRNAs), and 5′-NNNNGTAC-3′ PAM (yellow; 8 sgRNAs) were designed and their activities examined in HEK293 cells with deep sequencing. Error bars indicate s.e.m. (_n_=3).
Figure 3. Genome-wide target specificities of CjCas9 nucleases examined using Digenome-seq.
Human or mouse genomic DNA isolated from HeLa cells or NIH 3T3 cells (grey), respectively, was digested in vitro by Cas9 and its sgRNA targeted to the human AAVS1 locus (a,b,f) and the mouse Rosa26 (c), Vegfa (c,d) and Hif1a loci (d) and subjected to whole-genome sequencing. Circos plots show genome-wide DNA cleavage scores across the human or mouse genome. Red arrows indicate on-target sites. N indicates the number of in vitro cleavage sites identified by Digenome-seq. (a,b) Sequence logos were obtained by comparing DNA sequences at in vitro cleavage sites with each other. (b) Indel frequencies at the _AAVS1_-TS8 site measured using targeted deep sequencing. CjCas9 (orange) or SpCas9 (blue) targeted to the _AAVS1_-TS8 site was transfected into human HEK293 cells. Error bars indicate s.e.m. (_n_=3). (e) Indel frequencies at three AAVS1 sites targeted by CjCas9 (orange) and SaCas9 (violet).
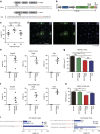
Figure 4. AAV-mediated mutagenesis in vitro and in vivo.
(a) AAV vector encoding CjCas9 and its sgRNA. (b) Indel frequencies at the Rosa26 target site in mouse C2C12 myotubes infected with AAV-CjCas9. (c) Indel frequencies at the Rosa26 target site in TA muscles of C57BL6J mice injected with AAV-CjCas9 measured at 2, 4, 8 and 32 weeks after injection. One-way ANOVA and Tukey's post hoc tests, **P<0.01, NS, not significant. See also Supplementary Fig. 3. (d) No off-target indels were detected at 20 homologous sites that differed from the on-target site by up to 4 nucleotides in the mouse genome. Genomic DNA isolated from AAV-CjCas9-injected TA muscles of C57BL6J mice mice at 32 weeks after injection was analysed by targeted deep sequencing. Mismatched nucleotides are shown in blue and PAM sequences in red. Red arrows indicate cleavage positions within the 20-bp target sequences. Error bars indicate s.e.m. (_n_=3).
Figure 5. In vivo genome editing with CjCas9 in the retina and retinal pigment epithelium.
(a) The CjCas9 target sequences in Vegfa and Hif1a/HIF1A genes. The PAM sequence and the sgRNA target sequence are shown in red and blue, respectively. (b) All-in-one AAV vector encoding CjCas9. (c) Indel frequencies at the Vegfa target site were analysed in RPE cells using deep sequencing at day 14, 28 and 42 post-intravitreal injection of AAV-CjCas9: Vegfa. Error bars indicate s.e.m. (_n_=4–5). One-way ANOVA and Tukey's post hoc tests, NS, not significant. (d) Representative confocal images of in vivo eGFP expression in RPE cells of AAV-CjCas9-injected mice 6 weeks after injection (_n_=6). eGFP was stained with anti-GFP antibody (green). Nuclei were counter-stained with DAPI (blue). Scale bar, 20 μm. (e–i) At day 42 post injection of AAV-CjCas9, indel frequencies and Vegfa protein levels were measured in retina and RPE cells using deep sequencing and ELISA, respectively. (e,f) Indel frequencies at the Rosa26, Vegfa and Hif1a target sites in the retina (e) and RPE cells (f). Error bars indicate s.e.m. (_n_=4 for AAV-uninjected control, _n_=5 for AAV-CjCas9). Student's _t_-tests, *P<0.05, ***P<0.001. (g,h) VEGFA levels measured by ELISA in the retina (g) and RPE cells (h), respectively. Error bars indicate s.e.m. (_n_=6–7). One-way ANOVA and Tukey's post hoc tests, *P<0.05, ***P<0.001. (i) Indel frequencies at in vitro cleavage sites identified by Digenome-seq. Genomic DNA isolated from RPE cells treated with AAV-CjCas9 at 6 weeks post injection was subjected to targeted deep sequencing. Mismatched nucleotides are shown in blue and PAM sequences in red. Red arrows indicate cleavage positions within the 22-bp target sequences.
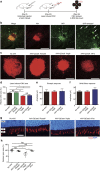
Figure 6. CjCas9 targeted to Vegfa or Hif1a reduces the area of laser-induced CNV in mice.
(a) At day 42 post injection of AAV-CjCas9, mice was treated with laser to induce choroidal neovascularization (CNV). One week after laser treatment, the CNV area was analysed. (b) In vivo expression of eGFP coexpressed with CjCas9 in laser-induced CNV. Representative confocal images of eGFP expression in the RPE of laser-induced CNV, 6 weeks after injection of AAV-CjCas9: Hif1a. eGFP was stained with anti-GFP antibody (green), CNV was stained with anti-IB4 antibody (red), and nuclei were counter-stained with DAPI (blue). Scale bar, 200 μm. Arrows indicate eGFP-expressing RPE cells. Scale bar, 100 μm (enlarged image). (c) Representative laser-induced CNV stained with isolectin B4 in the mouse eye injected with AAV-CjCas9 targeted to Rosa26, Vegfa or Hif1a. Scale bar, 200 μm. (d) The CNV area. Error bars indicate s.e.m. (_n_=17–18). One-way ANOVA and Tukey's post hoc tests, *P<0.05; **P<0.01; ***P<0.001; NS, not significant. (e,f) At day 56 post AAV injection, full-field electroretinogram (ERG) was performed to evaluate cone function in mice. There was no significant decrease of b-wave of photopic response (e) and 30 Hz flicker response (f) in both AAV-CjCas9: Vegfa (_n_=8) or AAV-CjCas9: Hif1a (_n_=6) treated mice, compared to normal control mice (_n_=8). Error bars indicate s.e.m. (_n_=6–8). (g,h) Opsin-positive areas in the retina at day 42 post injection. (g) Representative images of opsin-positive areas in contact with RPE cells expressing HA-tagged CjCas9 in AAV-CjCas9: Rosa26, Vegfa or Hif1a injected mice compared with the AAV-uninjected negative control mice (no AAV). Opsin (red) and DAPI (blue). Scale bar, 20 μm. ONL, outer nuclear layer; IS, inner segment of photoreceptor cells; OS, outer segment of photoreceptor cells. See also Supplementary Fig. 6. (h) Relative opsin areas of the AAV-CjCas9-injected mice were normalized to that of the AAV-uninjected negative control mice. Error bars indicate s.e.m. (_n_=4). One-way ANOVA and Tukey's post hoc tests, *P<0.05.
Similar articles
- Long-Term Effects of In Vivo Genome Editing in the Mouse Retina Using Campylobacter jejuni Cas9 Expressed via Adeno-Associated Virus.
Jo DH, Koo T, Cho CS, Kim JH, Kim JS, Kim JH. Jo DH, et al. Mol Ther. 2019 Jan 2;27(1):130-136. doi: 10.1016/j.ymthe.2018.10.009. Epub 2018 Oct 17. Mol Ther. 2019. PMID: 30470629 Free PMC article. - Targeted dual base editing with Campylobacter jejuni Cas9 by single AAV-mediated delivery.
Kweon J, Jang AH, Kwon E, Kim U, Shin HR, See J, Jang G, Lee C, Koo T, Kim S, Kim Y. Kweon J, et al. Exp Mol Med. 2023 Feb;55(2):377-384. doi: 10.1038/s12276-023-00938-w. Epub 2023 Feb 1. Exp Mol Med. 2023. PMID: 36720917 Free PMC article. - CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration.
Koo T, Park SW, Jo DH, Kim D, Kim JH, Cho HY, Kim J, Kim JH, Kim JS. Koo T, et al. Nat Commun. 2018 May 10;9(1):1855. doi: 10.1038/s41467-018-04175-y. Nat Commun. 2018. PMID: 29748595 Free PMC article. - Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox.
Karvelis T, Gasiunas G, Siksnys V. Karvelis T, et al. Curr Opin Microbiol. 2017 Jun;37:88-94. doi: 10.1016/j.mib.2017.05.009. Epub 2017 Jun 20. Curr Opin Microbiol. 2017. PMID: 28645099 Review. - Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment.
Kennedy EM, Cullen BR. Kennedy EM, et al. Virology. 2015 May;479-480:213-20. doi: 10.1016/j.virol.2015.02.024. Epub 2015 Mar 7. Virology. 2015. PMID: 25759096 Free PMC article. Review.
Cited by
- Harnessing the evolving CRISPR/Cas9 for precision oncology.
Li T, Li S, Kang Y, Zhou J, Yi M. Li T, et al. J Transl Med. 2024 Aug 8;22(1):749. doi: 10.1186/s12967-024-05570-4. J Transl Med. 2024. PMID: 39118151 Free PMC article. Review. - Developing small Cas9 hybrids using molecular modeling.
Mangin A, Dion V, Menzies G. Mangin A, et al. Sci Rep. 2024 Jul 26;14(1):17233. doi: 10.1038/s41598-024-68107-1. Sci Rep. 2024. PMID: 39060399 Free PMC article. - A most formidable arsenal: genetic technologies for building a better mouse.
Clark JF, Dinsmore CJ, Soriano P. Clark JF, et al. Genes Dev. 2020 Oct 1;34(19-20):1256-1286. doi: 10.1101/gad.342089.120. Genes Dev. 2020. PMID: 33004485 Free PMC article. Review. - CRISPR-Cas12a with an oAd Induces Precise and Cancer-Specific Genomic Reprogramming of EGFR and Efficient Tumor Regression.
Yoon AR, Jung BK, Choi E, Chung E, Hong J, Kim JS, Koo T, Yun CO. Yoon AR, et al. Mol Ther. 2020 Oct 7;28(10):2286-2296. doi: 10.1016/j.ymthe.2020.07.003. Epub 2020 Jul 3. Mol Ther. 2020. PMID: 32682455 Free PMC article. - Development and Application of CRISPR-Cas Based Tools.
Hu Y, Li W. Hu Y, et al. Front Cell Dev Biol. 2022 Apr 4;10:834646. doi: 10.3389/fcell.2022.834646. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35445018 Free PMC article. Review.
References
- Kim H. & Kim J. S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014). - PubMed
- Cho S. W., Kim S., Kim J. M. & Kim J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous