Gene expression profiling in the human alcoholic brain - PubMed (original) (raw)
Review
Gene expression profiling in the human alcoholic brain
Anna S Warden et al. Neuropharmacology. 2017.
Abstract
Long-term alcohol use causes widespread changes in gene expression in the human brain. Aberrant gene expression changes likely contribute to the progression from occasional alcohol use to alcohol use disorder (including alcohol dependence). Transcriptome studies have identified individual gene candidates that are linked to alcohol-dependence phenotypes. The use of bioinformatics techniques to examine expression datasets has provided novel systems-level approaches to transcriptome profiling in human postmortem brain. These analytical advances, along with recent developments in next-generation sequencing technology, have been instrumental in detecting both known and novel coding and non-coding RNAs, alternative splicing events, and cell-type specific changes that may contribute to alcohol-related pathologies. This review offers an integrated perspective on alcohol-responsive transcriptional changes in the human brain underlying the regulatory gene networks that contribute to alcohol dependence. This article is part of the Special Issue entitled "Alcoholism".
Keywords: Alcohol; Co-expression networks; Gene expression; RNA-seq; Transcriptional regulation.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
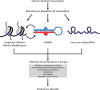
Figure 1
A hypothetical diagram for the role of transcriptional regulation in AUD. Chronic alcohol consumption can alter regulation of transcription via epigenetic modifications, including miRNAs and lncRNAs. Alterations in these regulatory mechanisms result in aberrant downstream gene expression changes in functional systems known to contribute to alcohol dependence (e.g. neuronal transmission, ion channels, immune signaling, stress response).
Figure 2
Total tissue versus cell-type specific analyses. Left Panel: Example volcano plot from an analysis of a cell-type specific isolation. The wider distribution of the plot signifies a greater number of statistically significant differentially expressed genes associated with chronic alcohol exposure. Right Panel: Example volcano plot from an analysis of a total tissue preparation. Note the smaller distribution and fewer statistically significant differentially expressed genes. Example data source: (
https://raw.githubusercontent.com/brennanpincardiff/RforBiochemists/master/data/microArrayData.tsv
”).
Figure 3
RNA-seq systems approach for understanding alcohol dependence. Sequencing can be used to evaluate changes in regulation of DNA and RNA expression in human alcoholic and control cases. Information from sequencing can then be superimposed with other variables such as protein data, physiological function, or other phenotypic information to identify mechanisms and potential treatment options for AUD. Adapted from (Farris and Mayfield, 2014).
References
- Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology. 1995;108:218–224. - PubMed
- Alaux-Cantin S, Warnault V, Legastelois R, Botia B, Pierrefiche O, Vilpoux C, Naassila M. Alcohol intoxications during adolescence increase motivation for alcohol in adult rats and induce neuroadaptations in the nucleus accumbens. Neuropharmacology. 2013;67:521–531. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 AA020683/AA/NIAAA NIH HHS/United States
- R01 AA012404/AA/NIAAA NIH HHS/United States
- R28 AA012725/AA/NIAAA NIH HHS/United States
- U01 AA020926/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources