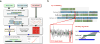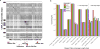Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome - PubMed (original) (raw)
. 2017 Apr;49(4):643-650.
doi: 10.1038/ng.3802. Epub 2017 Mar 6.
Benjamin D Rosen 2, Sergey Koren 3, Brian L Sayre 4, Alex R Hastie 5, Saki Chan 5, Joyce Lee 5, Ernest T Lam 5, Ivan Liachko 6, Shawn T Sullivan 7, Joshua N Burton 6, Heather J Huson 8, John C Nystrom 8, Christy M Kelley 9, Jana L Hutchison 2, Yang Zhou 2 10, Jiajie Sun 11, Alessandra Crisà 12, F Abel Ponce de León 13, John C Schwartz 14, John A Hammond 14, Geoffrey C Waldbieser 15, Steven G Schroeder 2, George E Liu 2, Maitreya J Dunham 6, Jay Shendure 6 16, Tad S Sonstegard 17, Adam M Phillippy 3, Curtis P Van Tassell 2, Timothy P L Smith 9
Affiliations
- PMID: 28263316
- PMCID: PMC5909822
- DOI: 10.1038/ng.3802
Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome
Derek M Bickhart et al. Nat Genet. 2017 Apr.
Abstract
The decrease in sequencing cost and increased sophistication of assembly algorithms for short-read platforms has resulted in a sharp increase in the number of species with genome assemblies. However, these assemblies are highly fragmented, with many gaps, ambiguities, and errors, impeding downstream applications. We demonstrate current state of the art for de novo assembly using the domestic goat (Capra hircus) based on long reads for contig formation, short reads for consensus validation, and scaffolding by optical and chromatin interaction mapping. These combined technologies produced what is, to our knowledge, the most continuous de novo mammalian assembly to date, with chromosome-length scaffolds and only 649 gaps. Our assembly represents a ∼400-fold improvement in continuity due to properly assembled gaps, compared to the previously published C. hircus assembly, and better resolves repetitive structures longer than 1 kb, representing the largest repeat family and immune gene complex yet produced for an individual of a ruminant species.
Conflict of interest statement
Competing Financial Interests: TSS is a current employee of Recombinetics. IL and STS are employees of Phase Genomics. JB, JS and MJD have a vested financial interest in Phase Genomics. ARH, SC, JL, and ETL are employees of Bionano Genomics. All other authors declare no competing financial interests.
Figures
Figure 1
Assembly schema for producing chromosome-length scaffolds. (A) Four different sets of sequencing data (long-read WGS, Hi-C data, optical mapping and short-read WGS) were produced in order to generate the goat reference genome. A tiered scaffolding approach using optical mapping data followed by Hi-C proximity guided assembly produced the highest quality genome assembly. (B) In order to correct misassemblies resulting from contig- or scaffold-errors, a consensus approach was used. An example from the initial optical mapping dataset is shown in the figure. A scaffold fork was identified on contig 3 (a 91 Mbp length contig) from the optical mapping data. Mapping of short-read WGS data showed signature that there was a misassembly near the 13th megabase of the contig, so it was split at this region. Subsequent analysis based on the RH map confirmed this split.
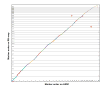
Figure 2
Assembly benchmarking comparisons reveal high degree of assembly completion. (A) Feature response curves (FRC) showing the error rate as a function of the number of bases in each assembly (CHIR_1.0, CHIR_2.0, and ARS1) and each scaffold test (intermediary assemblies using a combination of Hi-C and Bionano scaffolding). (B) Comparison plots of chromosome 20 sequence between the ARS1 and CHIR_2.0 assemblies reveal several small inversions (light blue circles) and a small insertion of sequence (break in continuity) in the ARS1 assembly. Red circles highlight 9 of the aforementioned inversions and the insertion of sequence in our assembly. The ARS1 assembly contains only 10 gaps on this chromosome scaffold whereas CHIR_2.0 has 5,651 gaps on the same chromosome assembly (gap density histogram on the Y axis). ARS1 optical map scaffolds and Pacbio contigs represented on the X axis as alternating patterns of blue and green shades, respectively, showing the tiling path that comprises the entire single chromosome scaffold.
Figure 3
RH probe map shows excellent assembly continuity. ARS1 RH probe mapping locations were plotted against the RH map order. Each ARS1 scaffold corresponds to an RH map chromosome with the exception of X which is composed of two scaffolds. Red circles highlight two intrachromosomal (on chrs 1 and 23) and two interchromosomal misassemblies (on chrs 18 and 17) in ARS1 that were difficult to resolve.
Figure 4
Long-read assembly with complementary scaffolding resolves gap regions (A) and long repeats (B) that cause problems for short-read reference annotation. (A) A region of the Mucin gene cluster was resolved by long-read assembly, resulting in a complete gene model for Mucin-5b-Like that was impossible due to two assembly gaps in the CHIR_2.0 assembly. (B) Counts of repetitive elements that had greater than 75% sequence length and greater than 60% identity with RepBase database entries for ruminant lineages. With the exception of the rRNA cluster (which is present in many repeated copies in the genome), the CHIR_2.0 reference contained a full complement of shorter repeat segments that were also present in our assembly. However, repeats that were larger than 1 kb were present in higher numbers in our assembly due to our ability to traverse the entire repetitive element’s length.
Figure 5
(A) A region of the Natural Killer Cell (NKC) gene cluster was fragmented in the CHIR_2.0 reference genome but is present on a single contig within ARS1. (B) Likewise, the Leukocyte Receptor Complex (LRC) locus was poorly represented in CHIR_2.0, and was missing ~500 kb of sequence. For highly repetitive and polymorphic gene families, our assembly approach provided the best resolution and highest continuity of gene sequence.
Similar articles
- Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity.
Low WY, Tearle R, Bickhart DM, Rosen BD, Kingan SB, Swale T, Thibaud-Nissen F, Murphy TD, Young R, Lefevre L, Hume DA, Collins A, Ajmone-Marsan P, Smith TPL, Williams JL. Low WY, et al. Nat Commun. 2019 Jan 16;10(1):260. doi: 10.1038/s41467-018-08260-0. Nat Commun. 2019. PMID: 30651564 Free PMC article. - De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture.
Mahajan S, Wei KH, Nalley MJ, Gibilisco L, Bachtrog D. Mahajan S, et al. PLoS Biol. 2018 Jul 30;16(7):e2006348. doi: 10.1371/journal.pbio.2006348. eCollection 2018 Jul. PLoS Biol. 2018. PMID: 30059545 Free PMC article. - Scaffolding of long read assemblies using long range contact information.
Ghurye J, Pop M, Koren S, Bickhart D, Chin CS. Ghurye J, et al. BMC Genomics. 2017 Jul 12;18(1):527. doi: 10.1186/s12864-017-3879-z. BMC Genomics. 2017. PMID: 28701198 Free PMC article. - The present and future of de novo whole-genome assembly.
Sohn JI, Nam JW. Sohn JI, et al. Brief Bioinform. 2018 Jan 1;19(1):23-40. doi: 10.1093/bib/bbw096. Brief Bioinform. 2018. PMID: 27742661 Review. - PacBio Sequencing and Its Applications.
Rhoads A, Au KF. Rhoads A, et al. Genomics Proteomics Bioinformatics. 2015 Oct;13(5):278-89. doi: 10.1016/j.gpb.2015.08.002. Epub 2015 Nov 2. Genomics Proteomics Bioinformatics. 2015. PMID: 26542840 Free PMC article. Review.
Cited by
- HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads.
Nurk S, Walenz BP, Rhie A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S. Nurk S, et al. Genome Res. 2020 Sep;30(9):1291-1305. doi: 10.1101/gr.263566.120. Epub 2020 Aug 14. Genome Res. 2020. PMID: 32801147 Free PMC article. - Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus).
Salgado Pardo JI, Delgado Bermejo JV, González Ariza A, León Jurado JM, Marín Navas C, Iglesias Pastrana C, Martínez Martínez MDA, Navas González FJ. Salgado Pardo JI, et al. Animals (Basel). 2022 Apr 11;12(8):988. doi: 10.3390/ani12080988. Animals (Basel). 2022. PMID: 35454235 Free PMC article. Review. - Whole genome analysis of Black Bengal goat from Savar Goat Farm, Bangladesh.
Chowdhury SMZH, Nazir KHMNH, Hasan S, Kabir A, Mahmud MM, Robbani M, Tabassum T, Afroze T, Rahman A, Islam MR, Hossain M. Chowdhury SMZH, et al. BMC Res Notes. 2019 Oct 24;12(1):687. doi: 10.1186/s13104-019-4700-7. BMC Res Notes. 2019. PMID: 31651366 Free PMC article. - A chromosome-scale genome assembly of the false clownfish, Amphiprion ocellaris.
Ryu T, Herrera M, Moore B, Izumiyama M, Kawai E, Laudet V, Ravasi T. Ryu T, et al. G3 (Bethesda). 2022 May 6;12(5):jkac074. doi: 10.1093/g3journal/jkac074. G3 (Bethesda). 2022. PMID: 35353192 Free PMC article. - Finding Nemo's Genes: A chromosome-scale reference assembly of the genome of the orange clownfish Amphiprion percula.
Lehmann R, Lightfoot DJ, Schunter C, Michell CT, Ohyanagi H, Mineta K, Foret S, Berumen ML, Miller DJ, Aranda M, Gojobori T, Munday PL, Ravasi T. Lehmann R, et al. Mol Ecol Resour. 2019 May;19(3):570-585. doi: 10.1111/1755-0998.12939. Epub 2018 Sep 10. Mol Ecol Resour. 2019. PMID: 30203521 Free PMC article.
References
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
- Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources