Autophagy and Microglia: Novel Partners in Neurodegeneration and Aging - PubMed (original) (raw)
Review
Autophagy and Microglia: Novel Partners in Neurodegeneration and Aging
Ainhoa Plaza-Zabala et al. Int J Mol Sci. 2017.
Abstract
Autophagy is emerging as a core regulator of Central Nervous System (CNS) aging and neurodegeneration. In the brain, it has mostly been studied in neurons, where the delivery of toxic molecules and organelles to the lysosome by autophagy is crucial for neuronal health and survival. However, we propose that the (dys)regulation of autophagy in microglia also affects innate immune functions such as phagocytosis and inflammation, which in turn contribute to the pathophysiology of aging and neurodegenerative diseases. Herein, we first describe the basic concepts of autophagy and its regulation, discuss key aspects for its accurate monitoring at the experimental level, and summarize the evidence linking autophagy impairment to CNS senescence and disease. We focus on acute, chronic, and autoimmunity-mediated neurodegeneration, including ischemia/stroke, Alzheimer's, Parkinson's, and Huntington's diseases, and multiple sclerosis. Next, we describe the actual and potential impact of autophagy on microglial phagocytic and inflammatory function. Thus, we provide evidence of how autophagy may affect microglial phagocytosis of apoptotic cells, amyloid-β, synaptic material, and myelin debris, and regulate the progression of age-associated neurodegenerative diseases. We also discuss data linking autophagy to the regulation of the microglial inflammatory phenotype, which is known to contribute to age-related brain dysfunction. Overall, we update the current knowledge of autophagy and microglia, and highlight as yet unexplored mechanisms whereby autophagy in microglia may contribute to CNS disease and senescence.
Keywords: aging; autophagy; inflammation; microglia; neurodegeneration; phagocytosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
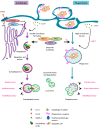
Figure 1
Autophagy and phagocytosis are lysosomal clearance pathways that share mechanistic and functional similarities. In response to cellular stress, autophagy (purple flow) is activated by signals that inhibit mechanistic target of rapamycin complex 1 (MTORC1) and activate unc-51 like autophagy activating kinase 1 (ULK-1), whereas phagocytosis (blue flow) is activated by extracellular ligands that bind to phagocytosis receptors in the surface of the microglial plasma membrane. Then, cargo engulfment structures start to form: the phagophore is de novo formed using the endoplasmic reticulum (ER) as a membrane source (autophagy) and the phagocytic cup is formed from invaginations of the plasma membrane (phagocytosis). These structures elongate and close up, forming the double-membrane-bound autophagosome (autophagy) and the single-membrane-containing phagosome (phagocytosis), which contain intracellular and extracellular degradative substrates, respectively. The formation of the autophagosome depends on the sequential and coordinated action of autophagy-related (ATGs) proteins, including microtubule-associated light chain 3 (LC3). In contrast, the formation of the phagosome may depend on the recruitment of autophagy machinery (ATGs and LC3) during LC3-associated phagocytosis (LAP) (described in peripheral macrophages, but not microglia; red question mark in the figure), or may be completed independently of ATGs in other types of phagocytosis. Finally, the autophagosome (autophagy) and the phagosome (phagocytosis), which contain the degradative cargo on their lumen, progressively mature and fuse with lysosomes, forming the autophagolysosome and the phagolysosome, respectively, where the cargo is finally digested. Autophagy and phagocytosis may serve similar functions in microglia, including the supply of energy during nutrient shortage, maintenance of cellular and tissue homeostasis, and the promotion of a net anti-inflammatory phenotype (see main text for details).
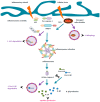
Figure 2
Autophagy may negatively regulate microglial inflammation: potential mechanisms of inflammasome regulation. Inflammasomes are cytosolic macromolecular sensors that assemble after activation by infectious or damaging stimuli (illustrated by blue arrows). They consist of a ligand sensor (i.e., NLRP3), an adaptor protein (ASC), and the immature form of the inflammatory caspase, pro-caspase-1. Inflammasome assembly induces the proteolytic processing of pro-caspase-1 to its active form caspase-1. Subsequently, activated caspase-1 proteolytically processes the immature forms of inflammatory cytokines pro-IL-1β and pro-IL-18 to active inflammatory mediators IL-1β and IL-18. In peripheral macrophages, three types of modulatory interactions have been described to explain the suppresive effect of autophagy over the inflammasome (illustrated by purple arrows). Thus, autophagy may target (1) the inflammasome adaptor protein ASC and/or (2) the inflammasome substrate pro-IL-1β for digestion to the lysosome. On the other hand, autophagy may (3) selectively digest damaged, ROS-generating mitochondria by mitophagy. Note that none of these mechanisms have yet been described in microglia (see main text for details).
Similar articles
- The effect of aged microglia on synaptic impairment and its relevance in neurodegenerative diseases.
Triviño JJ, von Bernhardi R. Triviño JJ, et al. Neurochem Int. 2021 Mar;144:104982. doi: 10.1016/j.neuint.2021.104982. Epub 2021 Feb 5. Neurochem Int. 2021. PMID: 33556444 Review. - Microglia and Aging: The Role of the TREM2-DAP12 and CX3CL1-CX3CR1 Axes.
Mecca C, Giambanco I, Donato R, Arcuri C. Mecca C, et al. Int J Mol Sci. 2018 Jan 22;19(1):318. doi: 10.3390/ijms19010318. Int J Mol Sci. 2018. PMID: 29361745 Free PMC article. Review. - Microglial Turnover in Ageing-Related Neurodegeneration: Therapeutic Avenue to Intervene in Disease Progression.
Azam S, Haque ME, Kim IS, Choi DK. Azam S, et al. Cells. 2021 Jan 14;10(1):150. doi: 10.3390/cells10010150. Cells. 2021. PMID: 33466587 Free PMC article. Review. - Impact of Aging in Microglia-Mediated D-Serine Balance in the CNS.
Beltrán-Castillo S, Eugenín J, von Bernhardi R. Beltrán-Castillo S, et al. Mediators Inflamm. 2018 Sep 27;2018:7219732. doi: 10.1155/2018/7219732. eCollection 2018. Mediators Inflamm. 2018. PMID: 30363571 Free PMC article. Review. - Lysosomal acidification dysfunction in microglia: an emerging pathogenic mechanism of neuroinflammation and neurodegeneration.
Quick JD, Silva C, Wong JH, Lim KL, Reynolds R, Barron AM, Zeng J, Lo CH. Quick JD, et al. J Neuroinflammation. 2023 Aug 5;20(1):185. doi: 10.1186/s12974-023-02866-y. J Neuroinflammation. 2023. PMID: 37543564 Free PMC article. Review.
Cited by
- Glial Cells in Glaucoma: Friends, Foes, and Potential Therapeutic Targets.
García-Bermúdez MY, Freude KK, Mouhammad ZA, van Wijngaarden P, Martin KK, Kolko M. García-Bermúdez MY, et al. Front Neurol. 2021 Mar 16;12:624983. doi: 10.3389/fneur.2021.624983. eCollection 2021. Front Neurol. 2021. PMID: 33796062 Free PMC article. Review. - Lysosomal Functions in Glia Associated with Neurodegeneration.
Kreher C, Favret J, Maulik M, Shin D. Kreher C, et al. Biomolecules. 2021 Mar 9;11(3):400. doi: 10.3390/biom11030400. Biomolecules. 2021. PMID: 33803137 Free PMC article. Review. - The innate immune response in tauopathies.
Johnson AM, Lukens JR. Johnson AM, et al. Eur J Immunol. 2023 Jun;53(6):e2250266. doi: 10.1002/eji.202250266. Epub 2023 May 4. Eur J Immunol. 2023. PMID: 36932726 Free PMC article. Review. - Understanding Epigenetics in the Neurodegeneration of Alzheimer's Disease: SAMP8 Mouse Model.
Griñán-Ferré C, Corpas R, Puigoriol-Illamola D, Palomera-Ávalos V, Sanfeliu C, Pallàs M. Griñán-Ferré C, et al. J Alzheimers Dis. 2018;62(3):943-963. doi: 10.3233/JAD-170664. J Alzheimers Dis. 2018. PMID: 29562529 Free PMC article. Review. - Autophagy in Neuroinflammation: A Focus on Epigenetic Regulation.
Chen Y, Chen J, Xing Z, Peng C, Li D. Chen Y, et al. Aging Dis. 2024 Apr 1;15(2):739-754. doi: 10.14336/AD.2023.0718-1. Aging Dis. 2024. PMID: 37548945 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical