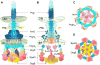In Situ Molecular Architecture of the Salmonella Type III Secretion Machine - PubMed (original) (raw)
In Situ Molecular Architecture of the Salmonella Type III Secretion Machine
Bo Hu et al. Cell. 2017.
Abstract
Type III protein secretion systems have specifically evolved to deliver bacterially encoded proteins into target eukaryotic cells. The core elements of this multi-protein machine are the envelope-associated needle complex, the inner membrane export apparatus, and a large cytoplasmic sorting platform. Here, we report a high-resolution in situ structure of the Salmonella Typhimurium type III secretion machine obtained by high-throughput cryo-electron tomography and sub-tomogram averaging. Through molecular modeling and comparative analysis of machines assembled with protein-tagged components or from different deletion mutants, we determined the molecular architecture of the secretion machine in situ and localized its structural components. We also show that docking of the sorting platform results in significant conformational changes in the needle complex to provide the symmetry adaptation required for the assembly of the entire secretion machine. These studies provide major insight into the structure and assembly of a broadly distributed protein secretion machine.
Keywords: Salmonella pathogenesis; bacterial pathogenesis; cryo electron tomography; molecular machines; protein secretion.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
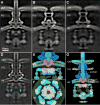
Figure 1. High resolution in situ structure of the entire type III protein secretion machine revealed by cryo-ET
(A) A schematic diagram of Salmonella delivering effector proteins into a target cell via a T3SS machine. (B) A schematic representation of the intact T3SS machine. (C) and (D) A tomographic slice (C) and a 3-D surface view (D) from a representative Salmonella minicell showing multiple injectisomes embedded in the cell envelope. (E and F) Central sections through different planes of the cytoplasmic sorting platform of a global average structure of the intact T3SS injectisome in situ show the components of the T3SS injectisome in the context of the outer membrane (OM), the peptidoglycan (PG), and the inner membrane (IM). (G–J) Cross-sections at the positions indicated in panel (E), are shown. See also Fig. S1, S4, and Table S1.
Figure 2. Comparison of the in situ and isolated structures of the type III secretion needle complex
(A) 3-D surface rendering of the intact injectisome structure shown in two different contour levels. Part of the map (colored in blue) matches well with the isolated NC structure. The rest of the map (green) can only be seen in situ. (B) 3D view of the superposition of the isolated NC protein density map (EMD-1875) onto the in situ injectisome structure. While the two structures match well in the central section (OR1, OR2 neck and IR1), a large shift of the IR2 is required to place it at its proper location in the cytoplasmic side of the inner membrane (IM) (C). The IR2 is placed within the inner membrane (B), to highlight a position that would be incompatible with the topology of the membrane protein components. (D) The atomic models of InvG and PrgH fit well into the 3D map of the intact injectisome in situ structure. See also Fig. S4 and Table S1.
Figure 3. Molecular architecture of the export apparatus in the intact T3SS machine
(A and B) A central section of the sub-tomogram average of the injectisome structure in wild type (A) and a Δ_invA_ deletion mutant (B). A large portion of the cytoplasmic complex remains, while a toroidal shape density immediately below the inner ring of the NC is absent. It appears that there is a ~5 nm fenestration (yellow arrow) in the inner leaflet of the inner membrane (B). (C) A central section of the sub-tomogram average of injectisomes from a quadruple deletion mutant lacking spaP, spaQ, spaR, and spaS. The toroidal shape density, the socket (orange arrow) and the needle fail to assemble, while a large portion of the cytopasmic complex remains intact. The fenestration observed at the inner membrane (B, yellow arrow) is no longer apparent in this mutant strain. Noticeably, the two leaflets of the inner membrane appear differently in the two mutants (B and C) and the wild type (A). (D) A central section of the sub-tomogram average of injectisomes from a strain expressing GFP-tagged InvA. Additional protein densities at the bottom the toroidal shape density (green arrows) are apparent in this strain, most likely representing the GFP tag added to the carboxy-terminus of InvA. (E–G) The nonameric ring atomic modeled structure of the carboxy-terminus of InvA fits well into the toroidal shape density in both side (E) and bottom (F) views. The additional densities assigned to GFP are shown in green (E and F). The InvA nonameric ring in the context of the entire injectisome is shown purple and the export apparatus in pink (G). See also Fig. S2, S3 and Table S1.
Figure 4. Structural characterization of the cytoplasmic sorting platform
Central sections of the sub-tomogram averages of injectisomes from Δ_orgA_ (A), Δ_orgB_ (B), Δ_spaO_ (C), Δ_invC_ (D), Δ_invi_ (E), and wild type (WT) (F) S. Typhimurium strains. The export apparatus appears to be intact in these mutants (A–E) compared to the WT injectisome structures (F). Most cytoplasmic densities are absent in Δ_orgA_ (A), Δ_orgB_ (B), and Δ_spaO_ (C), suggesting that these three proteins are essential for the assembly of the sorting platform. In the absence Δ_invC_ (D), the densities associated with the sorting platform are present although substantially reduced suggesting that InvC contributes to the stability of this structure. In contrast, in the Δ_invi_ mutant (E) the sorting platform is almost indistinguishable from wild type, suggesting a minor structural role for InvI. Central sections of the sub-tomogram averages of injectisomes obtained from strains expressing tagged versions of OrgA (G), OrgB (H), SpaO (i), InvC (J), and InvI (K) are shown. The location of the protein tag in the 3D rendering of the different structures is highlighted in panels as indicated (M–Q). A model of the proposed location of the different components of the sorting platform is shown in schematic (L) and 3D (R) manner (OrgA: green; InvC: orange; OrgB: yellow; SpaO: red; and PrgH: cyan. See also Fig. S2, S5, and Table S1.
Figure 5. Remodeling of PrgH amino-terminal domain upon sorting platform assembly
Organization of the amino-terminal cytoplasmic domain of PrgH (PrgHN) in the presence (A–E) or in the absence (F–J) of the sorting platform. Central (A and F) and cross (B and G) sections, or surface renderings (C and H) of the injectisomes from a S. Typhimurium wild type (A–C) or a strain lacking the sorting platform (Δ_orgA Δ_orgB Δ_spaO)_ (F–H) depicting the densities associated with PrgHN (light blue) are shown. The fitting of the atomic structure of PrgHN into the respective wild type (D and E) or Δ_orgA_ Δ_orgB_ Δ_spaO_ (I and J) injectisomes are shown depicting the conformational changes of PrgHN that occur upon assembly of the sorting platform. See also Fig. S5, S6, and Table S1.
Figure 6. Molecular model of the organization of the entire T3SS machine in situ
Side (A), cut–through (B), top (C), and bottom (D) views the intact injectisome structure. The available (or modeled) atomic structures of PrgHC, PrgHN, PrgK, PrgI, InvAC, OrgB, InvC and InvI have been fitted into the structure (see supplementary on line text). The location of the outer membrane (OM), inner membrane (IM), and peptidoglycan (PG) of the bacterial envelope are indicated.
Comment in
- Inside the Chamber of Secrets of the Type III Secretion System.
Cascales E. Cascales E. Cell. 2017 Mar 9;168(6):949-951. doi: 10.1016/j.cell.2017.02.028. Cell. 2017. PMID: 28283066
Similar articles
- Cryo-EM structure of the needle filament tip complex of the Salmonella type III secretion injectisome.
Guo EZ, Galán JE. Guo EZ, et al. Proc Natl Acad Sci U S A. 2021 Nov 2;118(44):e2114552118. doi: 10.1073/pnas.2114552118. Proc Natl Acad Sci U S A. 2021. PMID: 34706941 Free PMC article. - High-resolution view of the type III secretion export apparatus in situ reveals membrane remodeling and a secretion pathway.
Butan C, Lara-Tejero M, Li W, Liu J, Galán JE. Butan C, et al. Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24786-24795. doi: 10.1073/pnas.1916331116. Epub 2019 Nov 19. Proc Natl Acad Sci U S A. 2019. PMID: 31744874 Free PMC article. - A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion.
Guo EZ, Desrosiers DC, Zalesak J, Tolchard J, Berbon M, Habenstein B, Marlovits T, Loquet A, Galán JE. Guo EZ, et al. PLoS Biol. 2019 Jul 1;17(7):e3000351. doi: 10.1371/journal.pbio.3000351. eCollection 2019 Jul. PLoS Biol. 2019. PMID: 31260457 Free PMC article. - Assembly of the bacterial type III secretion machinery.
Diepold A, Wagner S. Diepold A, et al. FEMS Microbiol Rev. 2014 Jul;38(4):802-22. doi: 10.1111/1574-6976.12061. Epub 2014 Feb 17. FEMS Microbiol Rev. 2014. PMID: 24484471 Review. - Architecture and assembly of the Type VI secretion system.
Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. Zoued A, et al. Biochim Biophys Acta. 2014 Aug;1843(8):1664-73. doi: 10.1016/j.bbamcr.2014.03.018. Epub 2014 Mar 26. Biochim Biophys Acta. 2014. PMID: 24681160 Review.
Cited by
- Recent insights into type-3 secretion system injectisome structure and mechanism of human enteric pathogens.
Chen P, Goldberg MB. Chen P, et al. Curr Opin Microbiol. 2023 Feb;71:102232. doi: 10.1016/j.mib.2022.102232. Epub 2022 Nov 9. Curr Opin Microbiol. 2023. PMID: 36368294 Free PMC article. Review. - The Pseudomonas aeruginosa type III secretion translocator PopB assists the insertion of the PopD translocator into host cell membranes.
Tang Y, Romano FB, Breña M, Heuck AP. Tang Y, et al. J Biol Chem. 2018 Jun 8;293(23):8982-8993. doi: 10.1074/jbc.RA118.002766. Epub 2018 Apr 23. J Biol Chem. 2018. PMID: 29685888 Free PMC article. - Distinct Cytosolic Complexes Containing the Type III Secretion System ATPase Resolved by Three-Dimensional Single-Molecule Tracking in Live Yersinia enterocolitica.
Prindle JR, Wang Y, Rocha JM, Diepold A, Gahlmann A. Prindle JR, et al. Microbiol Spectr. 2022 Dec 21;10(6):e0174422. doi: 10.1128/spectrum.01744-22. Epub 2022 Nov 10. Microbiol Spectr. 2022. PMID: 36354362 Free PMC article. - Activation of Shigella flexneri type 3 secretion requires a host-induced conformational change to the translocon pore.
Russo BC, Duncan JK, Wiscovitch AL, Hachey AC, Goldberg MB. Russo BC, et al. PLoS Pathog. 2019 Nov 14;15(11):e1007928. doi: 10.1371/journal.ppat.1007928. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31725799 Free PMC article. - In Situ Molecular Architecture of the Helicobacter pylori Cag Type IV Secretion System.
Hu B, Khara P, Song L, Lin AS, Frick-Cheng AE, Harvey ML, Cover TL, Christie PJ. Hu B, et al. mBio. 2019 May 14;10(3):e00849-19. doi: 10.1128/mBio.00849-19. mBio. 2019. PMID: 31088930 Free PMC article.
References
- Agulleiro JI, Fernandez JJ. Tomo3D 2.0–exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol. 2015;189:147–152. - PubMed
- Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI112680/AI/NIAID NIH HHS/United States
- R37 AI030492/AI/NIAID NIH HHS/United States
- R01 AI087946/AI/NIAID NIH HHS/United States
- R01 GM107629/GM/NIGMS NIH HHS/United States
- R01 AI030492/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous