Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation - PubMed (original) (raw)
Canu: scalable and accurate long-read assembly via adaptive _k_-mer weighting and repeat separation
Sergey Koren et al. Genome Res. 2017 May.
Abstract
Long-read single-molecule sequencing has revolutionized de novo genome assembly and enabled the automated reconstruction of reference-quality genomes. However, given the relatively high error rates of such technologies, efficient and accurate assembly of large repeats and closely related haplotypes remains challenging. We address these issues with Canu, a successor of Celera Assembler that is specifically designed for noisy single-molecule sequences. Canu introduces support for nanopore sequencing, halves depth-of-coverage requirements, and improves assembly continuity while simultaneously reducing runtime by an order of magnitude on large genomes versus Celera Assembler 8.2. These advances result from new overlapping and assembly algorithms, including an adaptive overlapping strategy based on tf-idf weighted MinHash and a sparse assembly graph construction that avoids collapsing diverged repeats and haplotypes. We demonstrate that Canu can reliably assemble complete microbial genomes and near-complete eukaryotic chromosomes using either Pacific Biosciences (PacBio) or Oxford Nanopore technologies and achieves a contig NG50 of >21 Mbp on both human and Drosophila melanogaster PacBio data sets. For assembly structures that cannot be linearly represented, Canu provides graph-based assembly outputs in graphical fragment assembly (GFA) format for analysis or integration with complementary phasing and scaffolding techniques. The combination of such highly resolved assembly graphs with long-range scaffolding information promises the complete and automated assembly of complex genomes.
© 2017 Koren et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
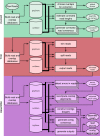
Figure 1.
A full Canu run includes three stages: correction (green), trimming (red), and assembly (purple). Canu stages share an interface for binary on-disk stores (databases), as well as parallel store construction. In all stages, the first step constructs an indexed store of input sequences, generates a _k_-mer histogram, constructs an indexed store of all-versus-all overlaps, and collates summary statistics. The correction stage (green) selects the best overlaps to use for correction, estimates corrected read lengths, and generates corrected reads. The trimming stage (red) identifies unsupported regions in the input and trims or splits reads to their longest supported range. The assembly stage (purple) makes a final pass to identify sequencing errors; constructs the best overlap graph (BOG); and outputs contigs, an assembly graph, and summary statistics.
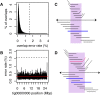
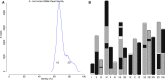
Figure 2.
An illustration of overlap error rate estimation, repeat identification, and splitting. (A) A histogram of all best edge error rates with the auto-selected threshold shown as a dashed line for the Drosophila melanogaster PacBio data set. All overlaps up to 4% error were computed. However, the modal error rate is 0.25% (0.25% median, 0.15% MAD), and Canu chose to use only overlaps <1.6% error for graph construction on this data set. (B) The dashed line shows the global error rate threshold (1.6%), and the profile shows the locally computed error rate for the largest contig in this assembly. Only overlaps consistent with this local error rate are considered as potential alternate paths when supplementing the initial BOG. By adjusting the error rate for each contig, Canu can separate diverged repeats without making an assumption of uniform read error across the assembly. (C) The contig is shown as a black line with arrows on both sides, indicating Bogart extends a path in both the 5′ and 3′ directions until encountering no overlaps or a read that is already incorporated in another contig. Repeat regions annotated by conflicting reads are shown above the contig. The reads align to part of the contig (the repeat) but indicate a different boundary sequence. A single read (blue line) spans the full repeat region, indicating the contig reconstruction is correct. (D) Repeat regions annotated by conflicting reads as before. In this case, no single read spans the full repeat region, and the initial contig was built using the overlap between two blue reads. The contig is split if the overlap between the two blue reads is not significantly better than the overlap from either blue read to the conflicting red read.
Figure 3.
Canu GFA output localizes complex repeat regions, allowing for improved scaffolding. (A) Bandage (Wick et al. 2015) plot of D. melanogaster compared with the karyotype (Stevens 1912; Metz 1914) from FlyBase (Attrill et al. 2016). Nodes are contigs sized by length, and edges indicate unused overlaps between contigs. The largest contigs are colored randomly and labeled with their chromosome based on alignment to the reference. (B) The callout shows Chromosome 2L from positions 3.07–23.12 Mbp, redrawn with the centromere at the top (indicated by a filled circle). Unique contigs are shaded black, while repeat contigs are shaded red. While the 2L chromosome scaffold is composed of 10 individual contigs, they are all linked in the output graph. The two red regions correspond to reference gaps at positions 2L:21,485,538, which consist of 100–200 copies of the histone gene cluster spanning >500 kbp and 2L:22,420,241, which is bordered by several TE repeats (Hoskins et al. 2015). The break in the bottom left of Chromosome 2L could not be confidently identified but is next to a feature labeled “FlyBase transposable element” in the genome annotation and is likely a transposable element insertion site. Even though Canu is unable to fully resolve these large repeat arrays, the graph indicates large-scale continuity across Chromosome 2L and could enable resolution with secondary technologies.
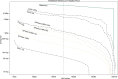
Figure 4.
A comparison of Arabidopsis thaliana assembly continuity for Canu and SPAdes. Each set of contigs is sorted from longest to shortest and plotted versus a cumulative percentage of the genome covered. Assemblies with larger contigs appear in the top of the plot. The ideal assembly corresponds to the green reference line. The commonly used NG50 metric corresponds to the vertical dashed line. Canu quickly gains continuity with increasing coverage, approaching the limit with 50× PacBio on this genome. In contrast, while making a large gain from Illumina-only to 10× PacBio, SPAdes continuity plateaus by 30×, and the Canu 20× assembly is comparable to the hybrid SPAdes assembly using 150× PacBio.
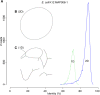
Figure 5.
Canu can assemble both 1D and 2D Nanopore Escherhicia coli reads. (A) A comparison of error rates for 1D and 2D read error rates versus the reference. Template 1D and 2D reads from the MAP006-1 E. coli data set were aligned independently to compute an identity for all reads with an alignment >90% of their length (95% of the 2D reads and 86% of the 1D reads had an alignment >90% of their length). The 2D sequences averaged 86% identity, and the 1D reads averaged 70% identity. (B) Bandage plot of the Canu BOG for the 2D data. The genome is in a single circle representing the full chromosome. (C) The corresponding plot for 1D data. While highly continuous, there are multiple components due to missed overlaps and unresolved repeats (due to the higher sequencing error rate).
Figure 6.
A highly continuous S. cerevisae assembly from noisy 1D and 2D MinION reads. (A) A histogram of read error rates (1D and 2D) versus the reference. Alignment identity was computed only for reads with an alignment >90% of their length. The majority of reads were <75% identity with an overall average of 70%. (_B_) Assembled Canu contigs were aligned to the reference, and all alignments >1 kbp in length and >90% identity were then plotted using the ColoredChromosomes package (Böhringer et al. 2002). Alternating shades indicate adjacent alignments, so each transition from gray to black represents a contig boundary or alignment breakpoint. White regions indicate regions missing from the assembly. Most chromosomes are in less than three contigs, indicating structural agreement with the reference.
Similar articles
- HINGE: long-read assembly achieves optimal repeat resolution.
Kamath GM, Shomorony I, Xia F, Courtade TA, Tse DN. Kamath GM, et al. Genome Res. 2017 May;27(5):747-756. doi: 10.1101/gr.216465.116. Epub 2017 Mar 20. Genome Res. 2017. PMID: 28320918 Free PMC article. - Improved assembly of noisy long reads by k-mer validation.
Carvalho AB, Dupim EG, Goldstein G. Carvalho AB, et al. Genome Res. 2016 Dec;26(12):1710-1720. doi: 10.1101/gr.209247.116. Epub 2016 Oct 7. Genome Res. 2016. PMID: 27831497 Free PMC article. - Fast and accurate de novo genome assembly from long uncorrected reads.
Vaser R, Sović I, Nagarajan N, Šikić M. Vaser R, et al. Genome Res. 2017 May;27(5):737-746. doi: 10.1101/gr.214270.116. Epub 2017 Jan 18. Genome Res. 2017. PMID: 28100585 Free PMC article. - One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly.
Koren S, Phillippy AM. Koren S, et al. Curr Opin Microbiol. 2015 Feb;23:110-20. doi: 10.1016/j.mib.2014.11.014. Epub 2014 Dec 1. Curr Opin Microbiol. 2015. PMID: 25461581 Review. - Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions.
Pucker B, Irisarri I, de Vries J, Xu B. Pucker B, et al. Quant Plant Biol. 2022 Mar 11;3:e5. doi: 10.1017/qpb.2021.18. eCollection 2022. Quant Plant Biol. 2022. PMID: 37077982 Free PMC article. Review.
Cited by
- Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing.
Bliznina A, Masunaga A, Mansfield MJ, Tan Y, Liu AW, West C, Rustagi T, Chien HC, Kumar S, Pichon J, Plessy C, Luscombe NM. Bliznina A, et al. BMC Genomics. 2021 Mar 29;22(1):222. doi: 10.1186/s12864-021-07512-6. BMC Genomics. 2021. PMID: 33781200 Free PMC article. - A novel fragmented mitochondrial genome in the protist pathogen Toxoplasma gondii and related tissue coccidia.
Namasivayam S, Baptista RP, Xiao W, Hall EM, Doggett JS, Troell K, Kissinger JC. Namasivayam S, et al. Genome Res. 2021 May;31(5):852-865. doi: 10.1101/gr.266403.120. Epub 2021 Apr 27. Genome Res. 2021. PMID: 33906963 Free PMC article. - Reevaluation of the Toxoplasma gondii and Neospora caninum genomes reveals misassembly, karyotype differences, and chromosomal rearrangements.
Berná L, Marquez P, Cabrera A, Greif G, Francia ME, Robello C. Berná L, et al. Genome Res. 2021 May;31(5):823-833. doi: 10.1101/gr.262832.120. Epub 2021 Apr 27. Genome Res. 2021. PMID: 33906964 Free PMC article. - Metagenomic Data Assembly - The Way of Decoding Unknown Microorganisms.
Lapidus AL, Korobeynikov AI. Lapidus AL, et al. Front Microbiol. 2021 Mar 23;12:613791. doi: 10.3389/fmicb.2021.613791. eCollection 2021. Front Microbiol. 2021. PMID: 33833738 Free PMC article. Review. - A chromosome-level assembly of the widely used Rockefeller strain of Aedes aegypti, the yellow fever mosquito.
Fisher CR, Wilson M, Scott JG. Fisher CR, et al. G3 (Bethesda). 2022 Nov 4;12(11):jkac242. doi: 10.1093/g3journal/jkac242. G3 (Bethesda). 2022. PMID: 36086997 Free PMC article.
References
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630. - PubMed
- Böhringer S, Gödde R, Böhringer D, Schulte T, Epplen JT. 2002. A software package for drawing ideograms automatically. Online J Bioinformatics 1: 51–61.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous