Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex - PubMed (original) (raw)
. 2017 Mar 23;169(1):47-57.e11.
doi: 10.1016/j.cell.2017.03.012.
Joshua Carter 2, MaryClare F Rollins 2, Sarah M Golden 2, Ryan N Jackson 2, Connor Hoffmann 2, Lyn'Al Nosaka 1, Joseph Bondy-Denomy 3, Karen L Maxwell 4, Alan R Davidson 5, Elizabeth R Fischer 6, Gabriel C Lander 7, Blake Wiedenheft 8
Affiliations
- PMID: 28340349
- PMCID: PMC5478280
- DOI: 10.1016/j.cell.2017.03.012
Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex
Saikat Chowdhury et al. Cell. 2017.
Abstract
Genetic conflict between viruses and their hosts drives evolution and genetic innovation. Prokaryotes evolved CRISPR-mediated adaptive immune systems for protection from viral infection, and viruses have evolved diverse anti-CRISPR (Acr) proteins that subvert these immune systems. The adaptive immune system in Pseudomonas aeruginosa (type I-F) relies on a 350 kDa CRISPR RNA (crRNA)-guided surveillance complex (Csy complex) to bind foreign DNA and recruit a trans-acting nuclease for target degradation. Here, we report the cryo-electron microscopy (cryo-EM) structure of the Csy complex bound to two different Acr proteins, AcrF1 and AcrF2, at an average resolution of 3.4 Å. The structure explains the molecular mechanism for immune system suppression, and structure-guided mutations show that the Acr proteins bind to residues essential for crRNA-mediated detection of DNA. Collectively, these data provide a snapshot of an ongoing molecular arms race between viral suppressors and the immune system they target.
Keywords: Acr; CRISPR; CRISPR-Cas; Cas; Csy; anti-CRISPRs; crRNA; cryo-EM; cryo-electron microscopy; type I-F.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
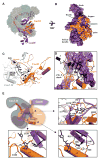
Figure 1. Structure of the Csy complex bound to two different virally encoded anti-CRISPR proteins
(A) The type I-F CRISPR-mediated immune system in P. aeruginosa (strain PA14) consists of six cas genes (legacy names are noted below each arrow) flanked by two CRISPR loci. The CRISPR loci are comprised of 28-nucleotide repeats (black diamonds) separated by 32-nucleotide phage or plasmid-derived spacer sequences (red cylinders) that lie downstream of an AT-rich leader sequence. (B) Schematic of Csy complex bound to two molecules of anti-CRISPR protein AcrF1 (red, F1.1 and F1.2) and one molecule of AcrF2 (green, F2). (C) Atomic model of the Csy complex (transparent pipes and planks) bound to AcrF1.1, AcrF1.2, and AcrF2. AcrF1 and F2 are shown as red and green surfaces, respectively. (D) Structures of the virally encoded anti-CRISPR proteins (Acr) and their locations in the complex. (E) Individual subunits of the Csy complex. The ‘thumb’ of each Cas7f subunit and Cas5f folds over the top of the crRNA, creating a kink in the RNA at 6-nt intervals (positions -1, 6, 12, 18, 24, and 30).
Figure 2. Structural similarities and differences between the Cas7-crRNA backbones of Class 1 CRISPR-Cas systems
(A) Comparison of type I-F (Csy), I-E (Cascade), and III-B (Cmr). The Cas7 backbone proteins (blue and grey) are shown as pipes and planks with head, tail, and belly proteins shown as transparent surfaces. (B) A single Cas7 homolog from each system (grey) bound to crRNA (red). Kinked bases formed by the thumbs are highlighted. (C) Nucleotides 1 to 5 (nt 1–5) of the crRNA-guide from each system were superimposed. The next six bases of each crRNA diverge after the kink. (D) Eight repeat-derived nucleotides on the 5′ end of the crRNA (black, also called the 5′-handle), were aligned using Chimera(Goddard et al., 2005). Differences in kink angles result in crRNA that have very different pitches. Expanded view highlights the conserved structure of the 5′-handle.
Figure 3. Assembly of the Cas5f/Cas8f tail through recognition of the 5′-handle
(A) Surface view of the Csy complex with Cas5f (orange) and Cas8f (purple) depicted as pipes and planks. (B) Surface representation of Cas5f (orange), showcasing the “left-handed fist” morphology and interactions with the Cas8f protein (purple). (C) Recognition of the S-shaped architecture of the 5′-handle by Cas7.6f (gray) and Cas5f (orange). (D) The 5′-end of the crRNA (black) is sandwiched between Cas8f harpoon and the first helix (α1) of the Cas5f RRM. (E) Cartoon depiction of Cas7.6f, Cas5f, Cas8f, and the 5′-end of the crRNA. Arrows point to detailed interactions between the Cas5f/Cas8f heterodimer and the crRNA at positions -6, -7, and -8.
Figure 4. Anti-CRISPR protein AcrF1 binds to residues on Cas7f that are essential for crRNA-guided recognition of target DNA
(A) Cartoon showing the location of three lysine residues (blue circles containing the letter K) on two adjacent Cas7f proteins that form a binding site for AcrF1. (B) Schematic of the Csy complex and the Acr proteins illustrating interactions between Acr and Cas proteins. The diameter of lines connecting subunits scales with buried surface. (C) Structure of two adjacent Cas7f proteins shown as gray (Cas7.6f) and cyan (Cas7.5f) surfaces. The three lysine residues (K85, K254, and K257) highlighted in panel A are shown in blue. AcrF1 is shown in dark red. Above is an expanded view of AcrF1.2 interacting with K85 on the thumb of Cas7.6f. Left is an expanded view of the acidic α2 from AcrF1.2 nestled against the positively charged residues on the extended web domains of Cas7.6f and Cas7.5f, respectively. Side chains involved in specific interactions are labeled. The disordered loop containing K257 is shown as a dotted C-α trace. (D) Surface plasmon resonance performed with wild type and mutant Csy complexes. Mutations in Cas7f (i.e. K85A or K254A/K257A) perturb AcrF1 binding. (E) AcrF1.1 binds to the thumb of Cas7.4f and AcrF1.2 binds to the thumb of Cas7.6f. In these two subunits (gray), but not the other four Cas7f proteins (cyan), the extended web is folded over the crRNA, restricting access to the guide. (F) Electrophoretic mobility shift assays performed with radiolabeled dsDNA substrates show that Cas7f K85A and K254A/K257A mutations perturb crRNA-guided DNA binding. Error bars, s.d.; n = 3.
Figure 5. Anti-CRISPR protein AcrF2 binds to a lysine-rich vise in the tail of Csy that is essential for DNA recognition
(A) Location of AcrF2 (green) in the complex with an expanded view of the AcrF2 binding site. Lysines on either Cas7.6f (i.e., K79 and K77) or Cas8f (i.e., K247, K28, and K31) that project into the AcrF2 binding site were mutated to glutamic acids. (B) Electrostatic representation of AcrF2 in the lysine-rich vise formed by Cas7.6f and Cas8f. (C) Surface representations of B-form dsDNA and AcrF2. Similarly positioned negative charges shown in red and orange. (D) Surface plasmon resonance performed with wild type and mutant Csy complexes. Mutations in Cas7f (i.e. Cas7f K77E/K79E) or Cas8f (Cas8f K247E, and Cas8f K28E/K31E) perturb AcrF2 binding. (E) Electrophoretic mobility shift assays were performed with radiolabeled dsDNA substrates. Mutations in the lysine-rich vise (Cas7K79E/K77E, Cas8K247E, or Cas8K28E/K31E) perturb binding to dsDNA targets. Error bars, s.d.; n = 3.
Similar articles
- Keeping crispr in check: diverse mechanisms of phage-encoded anti-crisprs.
Trasanidou D, Gerós AS, Mohanraju P, Nieuwenweg AC, Nobrega FL, Staals RHJ. Trasanidou D, et al. FEMS Microbiol Lett. 2019 May 1;366(9):fnz098. doi: 10.1093/femsle/fnz098. FEMS Microbiol Lett. 2019. PMID: 31077304 Free PMC article. Review. - Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures.
Peng R, Xu Y, Zhu T, Li N, Qi J, Chai Y, Wu M, Zhang X, Shi Y, Wang P, Wang J, Gao N, Gao GF. Peng R, et al. Cell Res. 2017 Jul;27(7):853-864. doi: 10.1038/cr.2017.79. Epub 2017 Jun 2. Cell Res. 2017. PMID: 28574055 Free PMC article. - Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Rollins MF, Chowdhury S, Carter J, Golden SM, Miettinen HM, Santiago-Frangos A, Faith D, Lawrence CM, Lander GC, Wiedenheft B. Rollins MF, et al. Mol Cell. 2019 Apr 4;74(1):132-142.e5. doi: 10.1016/j.molcel.2019.02.001. Epub 2019 Mar 11. Mol Cell. 2019. PMID: 30872121 Free PMC article. - CRISPR RNA and anti-CRISPR protein binding to the Xanthomonas albilineans Csy1-Csy2 heterodimer in the type I-F CRISPR-Cas system.
Hong S, Ka D, Yoon SJ, Suh N, Jeong M, Suh JY, Bae E. Hong S, et al. J Biol Chem. 2018 Feb 23;293(8):2744-2754. doi: 10.1074/jbc.RA117.001611. Epub 2018 Jan 18. J Biol Chem. 2018. PMID: 29348170 Free PMC article. - A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses.
Jackson RN, Wiedenheft B. Jackson RN, et al. Mol Cell. 2015 Jun 4;58(5):722-8. doi: 10.1016/j.molcel.2015.05.023. Epub 2015 May 28. Mol Cell. 2015. PMID: 26028539 Free PMC article. Review.
Cited by
- Current Updates of CRISPR/Cas System and Anti-CRISPR Proteins: Innovative Applications to Improve the Genome Editing Strategies.
Allemailem KS, Almatroudi A, Alrumaihi F, Alradhi AE, Theyab A, Algahtani M, Alhawas MO, Dobie G, Moawad AA, Rahmani AH, Khan AA. Allemailem KS, et al. Int J Nanomedicine. 2024 Oct 9;19:10185-10212. doi: 10.2147/IJN.S479068. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39399829 Free PMC article. Review. - Comparative genetic and genomic analysis of the novel fusellovirus Sulfolobus spindle-shaped virus 10.
Goodman DA, Stedman KM. Goodman DA, et al. Virus Evol. 2018 Aug 6;4(2):vey022. doi: 10.1093/ve/vey022. eCollection 2018 Jul. Virus Evol. 2018. PMID: 30094064 Free PMC article. - Keeping crispr in check: diverse mechanisms of phage-encoded anti-crisprs.
Trasanidou D, Gerós AS, Mohanraju P, Nieuwenweg AC, Nobrega FL, Staals RHJ. Trasanidou D, et al. FEMS Microbiol Lett. 2019 May 1;366(9):fnz098. doi: 10.1093/femsle/fnz098. FEMS Microbiol Lett. 2019. PMID: 31077304 Free PMC article. Review. - Anti-CRISPR: discovery, mechanism and function.
Pawluk A, Davidson AR, Maxwell KL. Pawluk A, et al. Nat Rev Microbiol. 2018 Jan;16(1):12-17. doi: 10.1038/nrmicro.2017.120. Epub 2017 Oct 24. Nat Rev Microbiol. 2018. PMID: 29062071 Review. - Guide RNA Categorization Enables Target Site Choice in Tn7-CRISPR-Cas Transposons.
Petassi MT, Hsieh SC, Peters JE. Petassi MT, et al. Cell. 2020 Dec 23;183(7):1757-1771.e18. doi: 10.1016/j.cell.2020.11.005. Epub 2020 Dec 2. Cell. 2020. PMID: 33271061 Free PMC article.
References
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. - PubMed
MeSH terms
Substances
Grants and funding
- DP5 OD021344/OD/NIH HHS/United States
- P30 GM110732/GM/NIGMS NIH HHS/United States
- F32 GM108436/GM/NIGMS NIH HHS/United States
- R01 GM110270/GM/NIGMS NIH HHS/United States
- P41 GM103310/GM/NIGMS NIH HHS/United States
- DP2 EB020402/EB/NIBIB NIH HHS/United States
- P20 GM103500/GM/NIGMS NIH HHS/United States
- R01 GM108888/GM/NIGMS NIH HHS/United States
- MOP-130482/CIHR/Canada
- MOP-136845/CIHR/Canada
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials