Gene and protein expressions and metabolomics exhibit activated redox signaling and wnt/β-catenin pathway are associated with metabolite dysfunction in patients with chronic kidney disease - PubMed (original) (raw)
Gene and protein expressions and metabolomics exhibit activated redox signaling and wnt/β-catenin pathway are associated with metabolite dysfunction in patients with chronic kidney disease
Dan-Qian Chen et al. Redox Biol. 2017 Aug.
Abstract
Changes in plasma concentration of small organic metabolites could be due to their altered production or urinary excretion and changes in their urine concentration may be due to the changes in their filtered load, tubular reabsorption, and/or altered urine volume. Therefore, these factors should be considered in interpretation of the changes observed in plasma or urine of the target metabolite(s). Fasting plasma and urine samples from 180 CKD patients and 120 age-matched healthy controls were determined by UPLC-HDMS-metabolomics and quantitative real-time RT-PCR techniques. Compared with healthy controls, patients with CKD showed activation of NF-κB and up-regulation of pro-inflammatory and pro-oxidant mRNA and protein expression as well as down-regulation of Nrf2-associated anti-oxidant gene mRNA and protein expression, accompanied by activated canonical Wnt/β-catenin signaling. 124 plasma and 128 urine metabolites were identified and 40 metabolites were significantly altered in both plasma and urine. Plasma concentration and urine excretion of 25 metabolites were distinctly different between CKD and controls. They were related to amino acid, methylamine, purine and lipid metabolisms. Logistic regression identified four plasma and five urine metabolites. Parts of them were good correlated with eGFR or serum creatinine. 5-Methoxytryptophan and homocystine and citrulline were good correlated with both eGFR and creatinine. Clinical factors were incorporated to establish predictive models. The enhanced metabolite model showed 5-methoxytryptophan, homocystine and citrulline have satisfactory accuracy, sensitivity and specificity for predictive CKD. The dysregulation of CKD was related to amino acid, methylamine, purine and lipid metabolisms. 5-methoxytryptophan, homocystine and citrulline could be considered as additional GFR-associated biomarker candidates and for indicating advanced renal injury. CKD caused dysregulation of the plasma and urine metabolome, activation of inflammatory/oxidative pathway and Wnt/β-catenin signaling and suppression of antioxidant pathway.
Keywords: Chronic kidney disease; Clinical factors; Inflammation; Metabolomics; Oxidative stress; Wnt/beta-catenin signaling.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
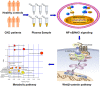
Graphical abstract
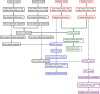
Fig. 1
Flow diagram of metabolomic analysis was used to show the overview of study design.
Fig. 2
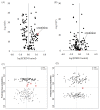
NF-κB target gene and protein expression in plasma from CKD patients. Quantitative real-time RT-PCR and Western blot depicting nuclear content of p65 active subunit of NF-κB and expression of COX-2, iNOS, MCP-1, P47Phox, p67Phox, Rac1 and gp91phox in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
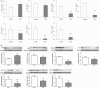
Fig. 3
Anti-oxidative stress Nrf2 and Nrf2 target gene and protein expression in plasma from CKD patients. Quantitative real-time RT-PCR and Western blot depicting Nrf2, catalase, HO-1, GPX, GCLC and NQO1 in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
Fig. 4
Total 16 Wnt and β-catenin gene expression as well as β-catenin and active β-catenin protein expression in plasma from CKD patients. Quantitative real-time RT-PCR including Wnt1, 2, 2b, 3, 4, 5a, 6, 7a, 7b, 8a, 8b, 9a, 9b, 10a, 10b and 16 and β-catenin as well as nuclear and cytoplasmic β-catenin and active β-catenin protein expression in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
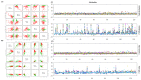
Fig. 5
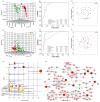
Metabolomic profiling of plasma and urine samples from two groups identifies metabolites that distinguish patients with CKD from controls. Outlier analysis of 124 identified metabolites in plasma (B) and 128 identified metabolites in urine (C). PCA of metabolites from 120 CKD samples and 80 control samples. Different principal components have a different contribution to separating CKD from controls in this study. Green crosses and red triangles represent controls and CKD, respectively. (D) Control-based z-score plot of metabolomic alterations from plasma and urine in control and patients with CKD. Each point represents an individual metabolite in one sample. Z-score plots for the data normalized to the mean of the control samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 6
The geometric mean ratio of each metabolite for individuals in CKD patients versus controls. The y-axis shows minus logarithm of P value. The x-axis shows the logarithm of ratio of CKD/control of each plasma sample (A) or urine sample (B). The log2(CKD/Control) with a value >0 indicated a relatively higher intensity present in CKD patients, whereas a value <0 indicated a relatively lower intensity compared with the healthy subjects. Diagnostic performances of the 25 metabolites in both plasma (D) and urine (E) based on the PLS-DA model. The black dots and black circles with red squares are for the incorrectly predicted samples in patient with CKD and controls, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 7
Multivariate analyses and correlation analysis of four significantly altered plasma metabolites in CKD patients. (A) The PCA score scatter plot using 27 differential metabolites from plasma sample between the CKD patients and controls. (B) Heatmap of 27 differential metabolites from plasma sample between the CKD patients and control. Red and blue in heatmap indicates increased and decreased levels, respectively. Rows: sample; columns: metabolite. THCA: trihydroxycoprostanoic acid. (C) PLS-DA-based ROC curves for the predictive power of four plasma biomarkers and for distinguishing CKD from controls. (D) Bar graphs of significant changes of four plasma biomarkers between CKD and controls. Abundance is represented as the relative intensity (y axis) of different groups (x axis). The statistical significance of differences between the two groups was marked. ***P<0.001 compared to the controls. (E) Correlation between canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine levels (peak intensity) measured by the UPLC-MS and eGFR by calculated formula. (F) Correlation between canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine levels (peak intensity) measured by the UPLC-MS and serum creatinine (µmol/L) measured by the clinical laboratory. The x-axes show the eGFR value or serum creatinine concentration. The y-axis shows the peak intensity of each plasma sample. The correlation coefficient is shown in each graph. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 8
Multivariate analyses and correlation analysis of five significantly altered urinary metabolites in CKD patients. (A) The PCA score scatter plot using 21 differential metabolites from urine sample between the CKD patients and controls. (B) Heatmap of 27 differential metabolites from urine sample between the CKD patients and control. Red and blue in heatmap indicates increased and decreased levels, respectively. Rows: sample; columns: metabolite. THCA: trihydroxycoprostanoic acid. (C) PLS-DA-based ROC curves for the predictive power of five urine metabolites and for distinguishing CKD from controls. (D) Bar graphs of significant changes of five urine metabolites between CKD and controls. Abundance is represented as the relative intensity (y axis) of different groups (x axis). The statistical significance of differences between the two groups was marked. ***P<0.001 compared to the controls. (E) Correlation between 1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline levels (peak intensity) measured by the UPLC-MS and eGFR by calculated formula. (F) Correlation between 1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline levels (peak intensity) measured by the UPLC-MS and serum creatinine (µmol/L) measured by the clinical laboratory. The x-axes show the eGFR value or serum creatinine concentration. The y-axis shows the peak intensity of each urine sample. The correlation coefficient is shown in each graph. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 9
Biomarker validations from four plasma metabolites and five urine metabolites from additional 60 CKD patients and 40 age-matched healthy controls. The three-dimensional PCA score scatter plot using four metabolites (canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine) in plasma (A) and five metabolites (1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline) in urine (D) between the patients with CKD and controls. PLS-DA-based ROC curves for the predictive power of four plasma biomarkers (B) and five urine biomarkers (E) for distinguishing CKD from controls. Diagnostic performance of the four plasma biomarkers (C) and five urine biomarkers (F) based on the PLS-DA model. The black dots or black circles with red squares are for the incorrectly predicted samples in patient with CKD and controls, respectively. (G) IPA with MetPA of 25 potential biomarkers combined plasma and urine. The size and color of each circle was based on pathway impact value and _p_-value, respectively. (H) Visualization of the remarkably disturbed metabolic pathways in plasma and urine by MetScape analysis. The identified metabolites in the current study were shown by red hexagons. Hexagons with green lines means that the significantly changes of the identified metabolite in CKD had statistical significance (P<0.05). The size of hexagons showed the fold change of the differential metabolite in CKD relative to control. In addition, pink hexagons showed metabolites participating in the metabolic pathway but not been identified in the current study. CKD were associated with purine metabolism, TCA cycle, aminoacyl-tRNA biosynthesis, nitrogen metabolism, taurine and hypotaurine metabolism, biotin metabolism, pantothenate and CoA biosynthesis, inositol phosphate metabolism, galactose metabolism, ascorbate and aldarate metabolism, primary bile acid biosynthesis, pyrimidine metabolism and steroid hormone biosynthesis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 10
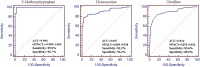
ROC curves analysis of enhanced PLS−DA model for the predictive power of combined each metabolite (5-MTP, homocystine and citrulline) and three general clinical parameters (age, gender and CKD vintage) for differentiating patients with CKD from controls.
Similar articles
- Activated NF-κB/Nrf2 and Wnt/β-catenin pathways are associated with lipid metabolism in CKD patients with microalbuminuria and macroalbuminuria.
Feng YL, Chen H, Chen DQ, Vaziri ND, Su W, Ma SX, Shang YQ, Mao JR, Yu XY, Zhang L, Guo Y, Zhao YY. Feng YL, et al. Biochim Biophys Acta Mol Basis Dis. 2019 Sep 1;1865(9):2317-2332. doi: 10.1016/j.bbadis.2019.05.010. Epub 2019 May 16. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 31102786 - The link between phenotype and fatty acid metabolism in advanced chronic kidney disease.
Chen DQ, Chen H, Chen L, Vaziri ND, Wang M, Li XR, Zhao YY. Chen DQ, et al. Nephrol Dial Transplant. 2017 Jul 1;32(7):1154-1166. doi: 10.1093/ndt/gfw415. Nephrol Dial Transplant. 2017. PMID: 28339984 - Plasma biomarker discovery for early chronic kidney disease diagnosis based on chemometric approaches using LC-QTOF targeted metabolomics data.
Benito S, Sánchez-Ortega A, Unceta N, Jansen JJ, Postma G, Andrade F, Aldámiz-Echevarria L, Buydens LMC, Goicolea MA, Barrio RJ. Benito S, et al. J Pharm Biomed Anal. 2018 Feb 5;149:46-56. doi: 10.1016/j.jpba.2017.10.036. Epub 2017 Oct 29. J Pharm Biomed Anal. 2018. PMID: 29100030 - Wnt/β-catenin signaling and renin-angiotensin system in chronic kidney disease.
Zhou L, Liu Y. Zhou L, et al. Curr Opin Nephrol Hypertens. 2016 Mar;25(2):100-6. doi: 10.1097/MNH.0000000000000205. Curr Opin Nephrol Hypertens. 2016. PMID: 26808707 Free PMC article. Review. - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
Cited by
- Mediterranean Diet Pattern: Potential Impact on the Different Altered Pathways Related to Cardiovascular Risk in Advanced Chronic Kidney Disease.
Rovira J, Ramirez-Bajo MJ, Bañon-Maneus E, Ventura-Aguiar P, Arias-Guillén M, Romano-Andrioni B, Ojeda R, Revuelta I, García-Calderó H, Barberà JA, Dantas AP, Diaz-Ricart M, Crispi F, García-Pagán JC, Campistol JM, Diekmann F. Rovira J, et al. Nutrients. 2024 Oct 31;16(21):3739. doi: 10.3390/nu16213739. Nutrients. 2024. PMID: 39519573 Free PMC article. - The Inhibitory Effects of Anti-GPC3 Antibody on Wnt/β-Catenin Signaling Pathway as a Biological Therapy in Liver Cancer.
Gan Q, Shao J, Sun T. Gan Q, et al. Mol Biotechnol. 2024 Sep 30. doi: 10.1007/s12033-024-01291-7. Online ahead of print. Mol Biotechnol. 2024. PMID: 39349719 - Causal effects of plasma metabolites on chronic kidney diseases and renal function: a bidirectional Mendelian randomization study.
Zhao X, Gao J, Kou K, Wang X, Gao X, Wang Y, Zhou H, Li F. Zhao X, et al. Front Endocrinol (Lausanne). 2024 Jul 26;15:1429159. doi: 10.3389/fendo.2024.1429159. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39129920 Free PMC article. - Interpretable machine learning identifies metabolites associated with glomerular filtration rate in type 2 diabetes patients.
An TF, Zhang ZP, Xue JT, Luo WM, Li Y, Fang ZZ, Zong GW. An TF, et al. Front Endocrinol (Lausanne). 2024 Jun 10;15:1279034. doi: 10.3389/fendo.2024.1279034. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38915893 Free PMC article. - Association of decreased estimated glomerular filtration rate with lung cancer risk in the Korean population.
Shin S, Kim MH, Oh CM, Chun H, Ha E, Lee HC, Moon SH, Lee DY, Cho D, Lee S, Jung MH, Ryoo JH. Shin S, et al. Epidemiol Health. 2024;46:e2024041. doi: 10.4178/epih.e2024041. Epub 2024 Mar 20. Epidemiol Health. 2024. PMID: 38549355 Free PMC article.
References
- Zhao Y. UPLC-based metabonomic applications for discovering biomarkers of diseases in clinical chemistry. Clin. Biochem. 2014;47(15):16–26. - PubMed
- Zhao Y. Intrarenal metabolomic investigation of chronic kidney disease and its TGF-β1 mechanism in induced-adenine rats using UPLC Q-TOF/HSMS/MSE. J. Proteome Res. 2013;12(2):692–703. - PubMed
- Zhang H. Metabolomic signatures of chronic kidney disease of diverse etiologies in the rats and humans. J. Proteome Res. 2016;15(10):3802–3812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous