Autophagy: machinery and regulation - PubMed (original) (raw)
Review
Autophagy: machinery and regulation
Zhangyuan Yin et al. Microb Cell. 2016.
Abstract
Macroautophagy/autophagy is an evolutionarily conserved cellular degradation process that targets cytoplasmic materials including cytosol, macromolecules and unwanted organelles. The discovery and analysis of autophagy-related (Atg) proteins have unveiled much of the machinery of autophagosome formation. Although initially autophagy was regarded as a survival response to stress, recent studies have revealed its significance in cellular and organismal homeostasis, development and immunity. Autophagic dysfunction and dysregulation are implicated in various diseases. In this review, we briefly summarize the physiological roles, molecular mechanism, regulatory network, and pathophysiological roles of autophagy.
Keywords: autophagosome formation; autophagy; cellular homeostasis; pathogenesis; physiological roles; regulation.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures
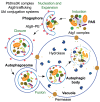
Figure 1. Figure 1: the mechanistic features of yeast autophagy.
Initiation of autophagy requires the formation of the Atg1 kinase complex at the PAS to allow the recruitment and activation of other Atg proteins. In yeast there is typically one PAS per cell, but the precise nature of this site is not well defined; the PAS may literally become a phagophore, making it a dynamic structure that continuously reforms. Next, the PtdIns3K complex translocates to the PAS to begin the nucleation process that will catalyze further movement of additional Atg proteins to this site. Ubl proteins such as Atg8–PE participate in cargo recognition during selective types of autophagy, and also play a role in determining the size of the autophagosome. Membrane delivery to the phagophore allows expansion and maturation into an autophagosome. This process also employs the transmembrane protein Atg9; however, the mechanism for phagophore expansion is poorly understood. Upon autophagosome completion, Atg4 deconjugates Atg8–PE on the surface of the autophagosome and the resulting vesicle fuses with the vacuole. In yeast, the inner vesicle is released into the lumen as an autophagic body. Within the vacuole, the contents of the autophagic body are released following lysis by the putative lipase Atg15. Finally, the cargo is degraded by resident hydrolases, and the resulting macromolecules are then released back into the cytosol via permeases.
Similar articles
- An overview of the molecular mechanism of autophagy.
Yang Z, Klionsky DJ. Yang Z, et al. Curr Top Microbiol Immunol. 2009;335:1-32. doi: 10.1007/978-3-642-00302-8_1. Curr Top Microbiol Immunol. 2009. PMID: 19802558 Free PMC article. Review. - An overview of macroautophagy in yeast.
Wen X, Klionsky DJ. Wen X, et al. J Mol Biol. 2016 May 8;428(9 Pt A):1681-99. doi: 10.1016/j.jmb.2016.02.021. Epub 2016 Feb 22. J Mol Biol. 2016. PMID: 26908221 Free PMC article. Review. - Autophagy core machinery: overcoming spatial barriers in neurons.
Ariosa AR, Klionsky DJ. Ariosa AR, et al. J Mol Med (Berl). 2016 Nov;94(11):1217-1227. doi: 10.1007/s00109-016-1461-9. Epub 2016 Aug 20. J Mol Med (Berl). 2016. PMID: 27544281 Free PMC article. Review. - Hidden behind autophagy: the unconventional roles of ATG proteins.
Bestebroer J, V'kovski P, Mauthe M, Reggiori F. Bestebroer J, et al. Traffic. 2013 Oct;14(10):1029-41. doi: 10.1111/tra.12091. Epub 2013 Jul 30. Traffic. 2013. PMID: 23837619 Free PMC article. Review. - Regulation of macroautophagy by mTOR and Beclin 1 complexes.
Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Pattingre S, et al. Biochimie. 2008 Feb;90(2):313-23. doi: 10.1016/j.biochi.2007.08.014. Epub 2007 Sep 8. Biochimie. 2008. PMID: 17928127 Review.
Cited by
- Canonical and Noncanonical Autophagy as Potential Targets for COVID-19.
Bello-Perez M, Sola I, Novoa B, Klionsky DJ, Falco A. Bello-Perez M, et al. Cells. 2020 Jul 5;9(7):1619. doi: 10.3390/cells9071619. Cells. 2020. PMID: 32635598 Free PMC article. Review. - Ablation of endothelial Atg7 inhibits ischemia-induced angiogenesis by upregulating Stat1 that suppresses Hif1a expression.
Yao H, Li J, Liu Z, Ouyang C, Qiu Y, Zheng X, Mu J, Xie Z. Yao H, et al. Autophagy. 2023 May;19(5):1491-1511. doi: 10.1080/15548627.2022.2139920. Epub 2022 Nov 21. Autophagy. 2023. PMID: 36300763 Free PMC article. - A Selective Look at Autophagy in Pancreatic β-Cells.
Pearson GL, Gingerich MA, Walker EM, Biden TJ, Soleimanpour SA. Pearson GL, et al. Diabetes. 2021 Jun;70(6):1229-1241. doi: 10.2337/dbi20-0014. Epub 2021 May 20. Diabetes. 2021. PMID: 34016598 Free PMC article. Review. - Herpud1 deficiency alleviates homocysteine-induced aortic valve calcification.
Xie W, Shan Y, Wu Z, Liu N, Yang J, Zhang H, Sun S, Chi J, Feng W, Lin H, Guo H. Xie W, et al. Cell Biol Toxicol. 2023 Dec;39(6):2665-2684. doi: 10.1007/s10565-023-09794-w. Epub 2023 Feb 6. Cell Biol Toxicol. 2023. PMID: 36746840 - Inhibition of phosphatidylinositol 3-kinase by PX-866 suppresses temozolomide-induced autophagy and promotes apoptosis in glioblastoma cells.
Harder BG, Peng S, Sereduk CP, Sodoma AM, Kitange GJ, Loftus JC, Sarkaria JN, Tran NL. Harder BG, et al. Mol Med. 2019 Nov 14;25(1):49. doi: 10.1186/s10020-019-0116-z. Mol Med. 2019. PMID: 31726966 Free PMC article.
References
Publication types
Grants and funding
This work was supported by NIH grant GM053396 to DJK.
LinkOut - more resources
Full Text Sources
Other Literature Sources