Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations - PubMed (original) (raw)
Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations
Alicia R Martin et al. Am J Hum Genet. 2017.
Erratum in
- Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations.
Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE. Martin AR, et al. Am J Hum Genet. 2020 Oct 1;107(4):788-789. doi: 10.1016/j.ajhg.2020.08.020. Am J Hum Genet. 2020. PMID: 33007199 Free PMC article. No abstract available.
Abstract
The vast majority of genome-wide association studies (GWASs) are performed in Europeans, and their transferability to other populations is dependent on many factors (e.g., linkage disequilibrium, allele frequencies, genetic architecture). As medical genomics studies become increasingly large and diverse, gaining insights into population history and consequently the transferability of disease risk measurement is critical. Here, we disentangle recent population history in the widely used 1000 Genomes Project reference panel, with an emphasis on populations underrepresented in medical studies. To examine the transferability of single-ancestry GWASs, we used published summary statistics to calculate polygenic risk scores for eight well-studied phenotypes. We identify directional inconsistencies in all scores; for example, height is predicted to decrease with genetic distance from Europeans, despite robust anthropological evidence that West Africans are as tall as Europeans on average. To gain deeper quantitative insights into GWAS transferability, we developed a complex trait coalescent-based simulation framework considering effects of polygenicity, causal allele frequency divergence, and heritability. As expected, correlations between true and inferred risk are typically highest in the population from which summary statistics were derived. We demonstrate that scores inferred from European GWASs are biased by genetic drift in other populations even when choosing the same causal variants and that biases in any direction are possible and unpredictable. This work cautions that summarizing findings from large-scale GWASs may have limited portability to other populations using standard approaches and highlights the need for generalized risk prediction methods and the inclusion of more diverse individuals in medical genomics.
Keywords: 1000 Genomes Project; GWAS; admixed populations; complex trait genetics; local ancestry; polygenic risk scores; population genetics; statistical genetics; summary statistics.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
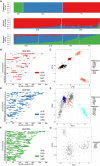
Sub-continental Diversity and Origins of African, European, and Native American Components of Recently Admixed American Populations (A) ADMIXTURE analysis at K = 3 focusing on admixed Americas samples, with the NAT, CEU, and YRI as reference populations. (B, D, and F) Local ancestry karyograms for representative PEL individual HG01893 with (B) African, (D) European, and (F) Native American components shown. (C, E, and G) Ancestry-specific PCA applied to admixed haploid genomes as well as ancestrally homogeneous continental reference populations from 1000 Genomes (where possible) for (C) African tracts, (E) European tracts, and (G) Native American tracts. A small number of admixed samples that constituted major outliers from the ancestry-specific PCA analysis were removed, including (C) one ASW sample (NA20314) and (E) eight samples, including three ACB, two ASW, one PEL, and two PUR samples.
Figure 2
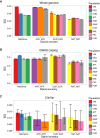
Heterozygosity by Continental and Diploid Local Ancestry Heterozygosity, estimated here as 2pq, is calculated in admixed populations stratified by diploid local ancestry in (A) the whole genome, (B) sites from the GWAS catalog, and (C) sites from ClinVar classified as “pathogenic” or “likely pathogenic.” The mean and 95% confidence intervals were calculated by bootstrapping 1,000 times. Populations not shown in a given panel have too few diploid ancestry tracts overlapping sites to calculate heterozygosity.
Figure 3
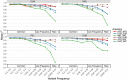
Imputation Accuracy by Local Ancestry in the Americas Accuracy was assessed via a leave-one-out strategy, stratified by diploid local ancestry on chromosome 9 for the Illumina OmniExpress genotyping array. Dashed lines indicate heterozygous diploid ancestry, and solid lines show homozygous diploid ancestry.
Figure 4
Biased Genetic Discoveries Influence Disease Risk Inferences Inferred and standardized polygenic risk scores across all individuals and colored by population for (A) height based on summary statistics from Wood et al., (B) schizophrenia based on summary statistics from the Schizophrenia Working Group of the Psychiatric Genomics Consortium, (C) type II diabetes summary statistics derived from a European cohort from Gaulton et al., and (D) type II diabetes summary statistics derived from a multiethnic cohort from Mahajan et al.
Figure 5
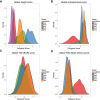
Coalescent Simulation Framework to Generate True and Inferred Polygenic Risk Scores Results of true and inferred polygenic risk scores, as well as their correlation, were computed via GWAS summary statistics from 10,000 simulated EUR case and control subjects modeling European, East Asian, and African population history (demographic parameters are from Gravel et al.14). (A) The distribution of mean true, unstandardized polygenic risk scores for each population across 500 simulations with m = 1,000 causal variants and _h_2 = 0.67. (B) The distribution of mean inferred, unstandardized polygenic risk for the same simulation parameters as in (A) (center) and standardized true versus inferred polygenic risk scores for three different coalescent simulation replicates showing 10,000 randomly drawn samples from each population not included as case or control subjects (right). (C) Violin plots show Pearson’s correlation across 50 iterations per parameter set between true and inferred polygenic risk scores across differing genetic architectures, including m = 200, 500, and 1,000 causal variants and _h_2 = 0.67.
Similar articles
- Variable prediction accuracy of polygenic scores within an ancestry group.
Mostafavi H, Harpak A, Agarwal I, Conley D, Pritchard JK, Przeworski M. Mostafavi H, et al. Elife. 2020 Jan 30;9:e48376. doi: 10.7554/eLife.48376. Elife. 2020. PMID: 31999256 Free PMC article. - Localizing Components of Shared Transethnic Genetic Architecture of Complex Traits from GWAS Summary Data.
Shi H, Burch KS, Johnson R, Freund MK, Kichaev G, Mancuso N, Manuel AM, Dong N, Pasaniuc B. Shi H, et al. Am J Hum Genet. 2020 Jun 4;106(6):805-817. doi: 10.1016/j.ajhg.2020.04.012. Epub 2020 May 21. Am J Hum Genet. 2020. PMID: 32442408 Free PMC article. - Impact of cross-ancestry genetic architecture on GWASs in admixed populations.
Mester R, Hou K, Ding Y, Meeks G, Burch KS, Bhattacharya A, Henn BM, Pasaniuc B. Mester R, et al. Am J Hum Genet. 2023 Jun 1;110(6):927-939. doi: 10.1016/j.ajhg.2023.05.001. Epub 2023 May 23. Am J Hum Genet. 2023. PMID: 37224807 Free PMC article. - The omnigenic model and polygenic prediction of complex traits.
Mathieson I. Mathieson I. Am J Hum Genet. 2021 Sep 2;108(9):1558-1563. doi: 10.1016/j.ajhg.2021.07.003. Epub 2021 Jul 30. Am J Hum Genet. 2021. PMID: 34331855 Free PMC article. Review. - Admixture and Ancestry Inference from Ancient and Modern Samples through Measures of Population Genetic Drift.
Harris AM, DeGiorgio M. Harris AM, et al. Hum Biol. 2017 Jan;89(1):21-46. doi: 10.13110/humanbiology.89.1.02. Hum Biol. 2017. PMID: 29285965 Review.
Cited by
- Association of polygenic liabilities for schizophrenia and bipolar disorder with educational attainment and cognitive aging.
Wu CS, Hsu CL, Lin MC, Su MH, Lin YF, Chen CY, Hsiao PC, Pan YJ, Chen PC, Huang YT, Wang SH. Wu CS, et al. Transl Psychiatry. 2024 Nov 16;14(1):472. doi: 10.1038/s41398-024-03182-6. Transl Psychiatry. 2024. PMID: 39550361 Free PMC article. - Familial coaggregation and shared genetic influence between major depressive disorder and gynecological diseases.
Chen CY, Cheng CF, Chen PC, Wu CS, Lin MC, Su MH, Chang CY, Pan YJ, Huang YT, Fan CC, Wang SH. Chen CY, et al. Eur J Epidemiol. 2024 Nov 4. doi: 10.1007/s10654-024-01166-w. Online ahead of print. Eur J Epidemiol. 2024. PMID: 39495462 - Nested admixture during and after the Trans-Atlantic Slave Trade on the island of São Tomé.
Ciccarella M, Laurent R, Szpiech ZA, Patin E, Dessarps-Freichey F, Utgé J, Lémée L, Semo A, Rocha J, Verdu P. Ciccarella M, et al. bioRxiv [Preprint]. 2024 Oct 23:2024.10.21.619344. doi: 10.1101/2024.10.21.619344. bioRxiv. 2024. PMID: 39484499 Free PMC article. Preprint. - The importance of family-based sampling for biobanks.
Davies NM, Hemani G, Neiderhiser JM, Martin HC, Mills MC, Visscher PM, Yengo L, Young AS, Keller MC. Davies NM, et al. Nature. 2024 Oct;634(8035):795-803. doi: 10.1038/s41586-024-07721-5. Epub 2024 Oct 23. Nature. 2024. PMID: 39443775 - Associations between polygenic scores for cognitive and non-cognitive factors of educational attainment and measures of behavior, psychopathology, and neuroimaging in the adolescent brain cognitive development study.
Gorelik AJ, Paul SE, Miller AP, Baranger DAA, Lin S, Zhang W, Elsayed NM, Modi H, Addala P, Bijsterbosch J, Barch DM, Karcher NR, Hatoum AS, Agrawal A, Bogdan R, Johnson EC. Gorelik AJ, et al. Psychol Med. 2024 Oct 23;54(13):1-15. doi: 10.1017/S0033291724002174. Online ahead of print. Psychol Med. 2024. PMID: 39440454 Free PMC article.
References
- Need A.C., Goldstein D.B. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. - PubMed
- Carlson C.S., Matise T.C., North K.E., Haiman C.A., Fesinmeyer M.D., Buyske S., Schumacher F.R., Peters U., Franceschini N., Ritchie M.D., PAGE Consortium Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11:e1001661. - PMC - PubMed
MeSH terms
Grants and funding
- U01 HG007417/HG/NHGRI NIH HHS/United States
- U01 HG007419/HG/NHGRI NIH HHS/United States
- U01 HG005208/HG/NHGRI NIH HHS/United States
- R01 GM083606/GM/NIGMS NIH HHS/United States
- U01 HG009080/HG/NHGRI NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- T32 GM007790/GM/NIGMS NIH HHS/United States
- U01 MH094432/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials