Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients - PubMed (original) (raw)
. 2017 Jul 15;65(2):208-215.
doi: 10.1093/cid/cix270.
Mirjana Mirceta 3, Ryan R Wick 1, David J Edwards 1, Nicholas R Thomson 4, Richard A Strugnell 2, Nigel F Pratt 5, Jill S Garlick 5, Kerri M Watson 5, David V Pilcher 6 7, Steve A McGloughlin 6 7, Denis W Spelman 8, Adam W J Jenney 8, Kathryn E Holt 1
Affiliations
- PMID: 28369261
- PMCID: PMC5850561
- DOI: 10.1093/cid/cix270
Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients
Claire L Gorrie et al. Clin Infect Dis. 2017.
Abstract
Background: Klebsiella pneumoniae is an opportunistic pathogen and leading cause of hospital-associated infections. Intensive care unit (ICU) patients are particularly at risk. Klebsiella pneumoniae is part of the healthy human microbiome, providing a potential reservoir for infection. However, the frequency of gut colonization and its contribution to infections are not well characterized.
Methods: We conducted a 1-year prospective cohort study in which 498 ICU patients were screened for rectal and throat carriage of K. pneumoniae shortly after admission. Klebsiella pneumoniae isolated from screening swabs and clinical diagnostic samples were characterized using whole genome sequencing and combined with epidemiological data to identify likely transmission events.
Results: Klebsiella pneumoniae carriage frequencies were estimated at 6% (95% confidence interval [CI], 3%-8%) among ICU patients admitted direct from the community, and 19% (95% CI, 14%-51%) among those with recent healthcare contact. Gut colonization on admission was significantly associated with subsequent infection (infection risk 16% vs 3%, odds ratio [OR] = 6.9, P < .001), and genome data indicated matching carriage and infection isolates in 80% of isolate pairs. Five likely transmission chains were identified, responsible for 12% of K. pneumoniae infections in ICU. In sum, 49% of K. pneumoniae infections were caused by the patients' own unique strain, and 48% of screened patients with infections were positive for prior colonization.
Conclusions: These data confirm K. pneumoniae colonization is a significant risk factor for infection in ICU, and indicate ~50% of K. pneumoniae infections result from patients' own microbiota. Screening for colonization on admission could limit risk of infection in the colonized patient and others.
Keywords: Klebsiella pneumoniae; gastrointestinal colonization; genomic epidemiology; hospital acquired infection; intensive care.
© The Author 2017. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
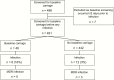
Figure 1.
Flowchart outlining number of patients included in each part of carriage rates analyses.
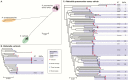
Figure 2.
Genome diversity of isolates from ICU patients identified as Klebsiella pneumoniae. All trees are maximum likelihood trees inferred from core genome SNP alignments. Scale bars indicate average number of substitutions per site across the genome. Tip colours indicate isolate source as per inset legend. *Possible mixed isolate (0.02–0.1 het/hom SNP ratio, excluded from pairwise SNP analysis in Figure 3). **Clinical isolate from sputum (KC0048), may not represent an infection. Phylogenetic lineages to which more than one ICU isolate belongs are highlighted and labeled with their corresponding multi-locus sequence type (ST) and the total number of SNPs identified between isolates in the lineage; darker shading indicates multiple patients contributed isolates in that cluster, as per inset legend. (A), Unrooted tree of all isolates, revealing three distinct species that are typically identified as K. pneumoniae in diagnostic laboratories. (B), Midpoint rooted species tree for K. variicola isolates. (C), Midpoint rooted species tree for K. pneumoniae sensu stricto isolates. Abbreviations: ICU, intensive care unit; SNP, single-nucleotide polymorphism.
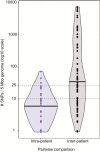
Figure 3.
Pairwise genetic distances between isolates belonging to the same lineage, expressed as SNPs per 5 Mbp of genome in order to normalise for differences in shared gene content between strain pairs. Violin plots showing distribution of pairwise genetic distances intra- and inter-patient; black bars indicate the median value. Note the log10 scale which excludes display of 1 strain pair that was separated by 0 SNPs. Abbreviation: SNP, single-nucleotide polymorphism.
Figure 4.
Timelines for all lineages detected in multiple patients that show any inter-patient pairwise genetic distance between isolates of ≤25 SNPs per 5 Mbp. Lineages are boxed and labeled with their multi-locus sequence type (ST). Each horizontal dashed line indicates the time line for a patient, labelled to the left (crosses indicate date of death where applicable). Periods of Alfred Hospital admission are indicated as white boxes, periods in ICU as pink shading. Circles indicate K. pneumoniae infection isolates (red, belonging to the lineage; black, other lineage); triangles indicate rectal screening swabs (red, K. pneumoniae belonging to the lineage; black, K. pneumoniae of another lineage; unfilled, negative for K. pneumoniae). Orange boxes indicate groups of isolates for which all patients have at least one pairwise genetic distance of ≤10 SNPs per 5 Mbp with another in the group; similarly for yellow (≤25 SNPs) and blue (≤100 SNPs) boxes. Abbreviations: ICU, intensive care unit; SNP, single-nucleotide polymorphism.
Comment in
- Genome watch: Klebsiella pneumoniae: when a colonizer turns bad.
Dorman MJ, Short FL. Dorman MJ, et al. Nat Rev Microbiol. 2017 Jul;15(7):384. doi: 10.1038/nrmicro.2017.64. Epub 2017 Jun 5. Nat Rev Microbiol. 2017. PMID: 28579608 No abstract available.
Similar articles
- The Colonization of Carbapenem-Resistant Klebsiella pneumoniae: Epidemiology, Resistance Mechanisms, and Risk Factors in Patients Admitted to Intensive Care Units in China.
Qin X, Wu S, Hao M, Zhu J, Ding B, Yang Y, Xu X, Wang M, Yang F, Hu F. Qin X, et al. J Infect Dis. 2020 Mar 16;221(Suppl 2):S206-S214. doi: 10.1093/infdis/jiz622. J Infect Dis. 2020. PMID: 32176790 - Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity.
Raffelsberger N, Hetland MAK, Svendsen K, Småbrekke L, Löhr IH, Andreassen LLE, Brisse S, Holt KE, Sundsfjord A, Samuelsen Ø, Gravningen K. Raffelsberger N, et al. Gut Microbes. 2021 Jan-Dec;13(1):1939599. doi: 10.1080/19490976.2021.1939599. Gut Microbes. 2021. PMID: 34182896 Free PMC article. - Spread of Carbapenem-Resistant Klebsiella pneumoniae in an Intensive Care Unit: A Whole-Genome Sequence-Based Prospective Observational Study.
Wei L, Wu L, Wen H, Feng Y, Zhu S, Liu Y, Tang L, Doughty E, van Schaik W, McNally A, Zong Z. Wei L, et al. Microbiol Spectr. 2021 Sep 3;9(1):e0005821. doi: 10.1128/Spectrum.00058-21. Epub 2021 Jul 14. Microbiol Spectr. 2021. PMID: 34259540 Free PMC article. - Deciphering the gastrointestinal carriage of Klebsiella pneumoniae.
Bray AS, Zafar MA. Bray AS, et al. Infect Immun. 2024 Sep 10;92(9):e0048223. doi: 10.1128/iai.00482-23. Epub 2024 Apr 10. Infect Immun. 2024. PMID: 38597634 Free PMC article. Review. - The correlation between intestinal colonization and infection of carbapenem-resistant Klebsiella pneumoniae: A systematic review.
Cai S, Wang Z, Han X, Hu H, Quan J, Jiang Y, Du X, Zhou Z, Yu Y. Cai S, et al. J Glob Antimicrob Resist. 2024 Sep;38:187-193. doi: 10.1016/j.jgar.2024.04.013. Epub 2024 May 21. J Glob Antimicrob Resist. 2024. PMID: 38777180
Cited by
- Animal Model To Study Klebsiella pneumoniae Gastrointestinal Colonization and Host-to-Host Transmission.
Young TM, Bray AS, Nagpal RK, Caudell DL, Yadav H, Zafar MA. Young TM, et al. Infect Immun. 2020 Oct 19;88(11):e00071-20. doi: 10.1128/IAI.00071-20. Print 2020 Oct 19. Infect Immun. 2020. PMID: 32839189 Free PMC article. - Carbapenem-resistant Acinetobacter baumannii: A challenge in the intensive care unit.
Jiang Y, Ding Y, Wei Y, Jian C, Liu J, Zeng Z. Jiang Y, et al. Front Microbiol. 2022 Nov 10;13:1045206. doi: 10.3389/fmicb.2022.1045206. eCollection 2022. Front Microbiol. 2022. PMID: 36439795 Free PMC article. Review. - Emerging Antimicrobial-Resistant High-Risk Klebsiella pneumoniae Clones ST307 and ST147.
Peirano G, Chen L, Kreiswirth BN, Pitout JDD. Peirano G, et al. Antimicrob Agents Chemother. 2020 Sep 21;64(10):e01148-20. doi: 10.1128/AAC.01148-20. Print 2020 Sep 21. Antimicrob Agents Chemother. 2020. PMID: 32747358 Free PMC article. Review. - Genomic investigation of the emergence of vanD vancomycin-resistant Enterococcus faecium.
Baines SL, Guérillot R, Ballard S, Johnson PDR, Stinear TP, Roberts S, Howden BP. Baines SL, et al. Access Microbiol. 2023 Dec 4;5(12):000712.v3. doi: 10.1099/acmi.0.000712.v3. eCollection 2023. Access Microbiol. 2023. PMID: 38188239 Free PMC article. - A curated collection of Klebsiella metabolic models reveals variable substrate usage and gene essentiality.
Hawkey J, Vezina B, Monk JM, Judd LM, Harshegyi T, López-Fernández S, Rodrigues C, Brisse S, Holt KE, Wyres KL. Hawkey J, et al. Genome Res. 2022 May;32(5):1004-1014. doi: 10.1101/gr.276289.121. Epub 2022 Mar 11. Genome Res. 2022. PMID: 35277433 Free PMC article.
References
- Centres for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013: Centres for Disease Control and Prevention, US Department of Health and Human Services, 2013.
- Paterson DL, Ko WC, Von Gottberg A, et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis 2004; 39:31–7. - PubMed
- Wiener-Well Y, Rudensky B, Yinnon AM, et al. Carriage rate of carbapenem-resistant Klebsiella pneumoniae in hospitalised patients during a national outbreak. J Hosp Infect 2010; 74:344–9. - PubMed
- Peña C, Pujol M, Ricart A, et al. Risk factors for faecal carriage of Klebsiella pneumoniae producing extended spectrum beta-lactamase (ESBL-KP) in the intensive care unit. J Hosp Infect 1997; 35:9–16. - PubMed
- Weinstein RA, Gaynes R, Edwards JR; National Nosocomial Infections Surveillance System Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 2005; 41:848–54. - PubMed