East African origins for Madagascan chickens as indicated by mitochondrial DNA - PubMed (original) (raw)
. 2017 Mar 22;4(3):160787.
doi: 10.1098/rsos.160787. eCollection 2017 Mar.
Affiliations
- PMID: 28405364
- PMCID: PMC5383821
- DOI: 10.1098/rsos.160787
East African origins for Madagascan chickens as indicated by mitochondrial DNA
Michael B Herrera et al. R Soc Open Sci. 2017.
Abstract
The colonization of Madagascar by Austronesian-speaking people during AD 50-500 represents the most westerly point of the greatest diaspora in prehistory. A range of economically important plants and animals may have accompanied the Austronesians. Domestic chickens (Gallus gallus) are found in Madagascar, but it is unclear how they arrived there. Did they accompany the initial Austronesian-speaking populations that reached Madagascar via the Indian Ocean or were they late arrivals with Arabian and African sea-farers? To address this question, we investigated the mitochondrial DNA control region diversity of modern chickens sampled from around the Indian Ocean rim (Southeast Asia, South Asia, the Arabian Peninsula, East Africa and Madagascar). In contrast to the linguistic and human genetic evidence indicating dual African and Southeast Asian ancestry of the Malagasy people, we find that chickens in Madagascar only share a common ancestor with East Africa, which together are genetically closer to South Asian chickens than to those in Southeast Asia. This suggests that the earliest expansion of Austronesian-speaking people across the Indian Ocean did not successfully introduce chickens to Madagascar. Our results further demonstrate the complexity of the translocation history of introduced domesticates in Madagascar.
Keywords: Madagascar; chicken; dispersal; human migration; mitochondrial DNA.
Figures
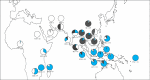
Figure 1.
Frequency distribution of chicken mitochondrial DNA haplogroup in the region under study (blue, haplogroup D; white, haplogroup E; grey, other haplogroups) with geographical location and sample size noted. Sample localities are Azerbaijan (AZR), Bangladesh (BLH), Burma (BUR), India (IND), Iran (IRA), Irian Jaya (IRJ), Java (JAV), Kalimantan (KAL), Kenya (KEN), Korea (KOR), Laos (LAO), Madagascar (MAD), Malawi (MLW), Maluku (MLK), Nigeria (NIG), Nusa Tenggara (NUS), Pacific (PAC; Fiji, Solomon and Vanuatu), Philippines (PHL; Luzon, Visayas and Mindanao), Saudi Arabia (SAU), South China (SC), Sri Lanka (SRI), Sudan (SUD), Sulawesi (SUL), Sumatra (SUM), Thailand (THA), Turkmenistan (TRK), Vietnam (VIE) and Zimbabwe (ZIM).
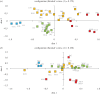
Figure 2.
Multidimensional scaling plots (MDS) for pairwise population Slatkin's linearized _F_ST for (a) 3128 chickens from Asia (orange), Africa (red), ISEA (green) and the Pacific (blue) using all haplogroups. (b) 1081 haplogroup D chickens from the same regions. Azerbaijan, Iran, Korea, Saudi Arabia, Sudan and Turkmenistan are not present in plot B because they do not contain haplogroup D lineages. See figure 1 for locality abbreviations.
Figure 3.
Median-joining network depicting the relationship of D haplotypes of chickens from East Africa and Madagascar (blue), South Asia (black) and Indonesia (green) using all observed D haplotypes regardless of frequency. Stars mark the position of the Madagascan samples. Inferred haplotypes are indicated by small red dots.
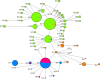
Figure 4.
Median-joining (MJ) network of mtDNA-CR D haplotypes observed in Madagascar (purple), Africa (blue), South Asia (brown) and Indonesia (green) excluding most haplotypes represented by one sample. The circle sizes are proportional to the haplotype frequencies and the length of the lines corresponds to the number of mutations connecting haplotypes.
Similar articles
- Origin and spatial population structure of Malagasy native chickens based on mitochondrial DNA.
Yonezawa T, Mannen H, Honma K, Matsunaga M, Rakotondraparany F, Ratsoavina FM, Wu J, Nishibori M, Yamamoto Y. Yonezawa T, et al. Sci Rep. 2024 Jan 4;14(1):569. doi: 10.1038/s41598-023-50708-x. Sci Rep. 2024. PMID: 38177203 Free PMC article. - Evidence of Austronesian Genetic Lineages in East Africa and South Arabia: Complex Dispersal from Madagascar and Southeast Asia.
Brucato N, Fernandes V, Kusuma P, Černý V, Mulligan CJ, Soares P, Rito T, Besse C, Boland A, Deleuze JF, Cox MP, Sudoyo H, Stoneking M, Pereira L, Ricaut FX. Brucato N, et al. Genome Biol Evol. 2019 Mar 1;11(3):748-758. doi: 10.1093/gbe/evz028. Genome Biol Evol. 2019. PMID: 30715341 Free PMC article. - African origin for Madagascan dogs revealed by mtDNA analysis.
Ardalan A, Oskarsson MC, van Asch B, Rabakonandriania E, Savolainen P. Ardalan A, et al. R Soc Open Sci. 2015 May 20;2(5):140552. doi: 10.1098/rsos.140552. eCollection 2015 May. R Soc Open Sci. 2015. PMID: 26064658 Free PMC article. - The History of African Village Chickens: an Archaeological and Molecular Perspective.
Mwacharo JM, Bjørnstad G, Han JL, Hanotte O. Mwacharo JM, et al. Afr Archaeol Rev. 2013;30(1):97-114. doi: 10.1007/s10437-013-9128-1. Epub 2013 Mar 3. Afr Archaeol Rev. 2013. PMID: 27212780 Free PMC article. Review. - Genetic evidence and historical theories of the Asian and African origins of the present Malagasy population.
Heiske M, Alva O, Pereda-Loth V, Van Schalkwyk M, Radimilahy C, Letellier T, Rakotarisoa JA, Pierron D. Heiske M, et al. Hum Mol Genet. 2021 Apr 26;30(R1):R72-R78. doi: 10.1093/hmg/ddab018. Hum Mol Genet. 2021. PMID: 33481023 Review.
Cited by
- Origin and spatial population structure of Malagasy native chickens based on mitochondrial DNA.
Yonezawa T, Mannen H, Honma K, Matsunaga M, Rakotondraparany F, Ratsoavina FM, Wu J, Nishibori M, Yamamoto Y. Yonezawa T, et al. Sci Rep. 2024 Jan 4;14(1):569. doi: 10.1038/s41598-023-50708-x. Sci Rep. 2024. PMID: 38177203 Free PMC article. - Modeling the Impact of Newcastle Disease Virus Vaccinations on Chicken Production Systems in Northeastern Madagascar.
Annapragada A, Borgerson C, Iams S, Ravelomanantsoa MA, Crawford GC, Helin M, Anjaranirina EJG, Randriamady HJ, Golden CD. Annapragada A, et al. Front Vet Sci. 2019 Sep 26;6:305. doi: 10.3389/fvets.2019.00305. eCollection 2019. Front Vet Sci. 2019. PMID: 31612142 Free PMC article. - European and Asian contribution to the genetic diversity of mainland South American chickens.
Herrera MB, Kraitsek S, Alcalde JA, Quiroz D, Revelo H, Alvarez LA, Rosario MF, Thomson V, Jianlin H, Austin JJ, Gongora J. Herrera MB, et al. R Soc Open Sci. 2020 Feb 5;7(2):191558. doi: 10.1098/rsos.191558. eCollection 2020 Feb. R Soc Open Sci. 2020. PMID: 32257320 Free PMC article. - Uganda chicken genetic resources: II. genetic diversity and population demographic history inferred from mitochondrial DNA D-loop sequences.
Yussif I, Kugonza DR, Masembe C. Yussif I, et al. Front Genet. 2024 Mar 7;15:1325569. doi: 10.3389/fgene.2024.1325569. eCollection 2024. Front Genet. 2024. PMID: 38516375 Free PMC article. - Population Genetic Structure and Contribution of Philippine Chickens to the Pacific Chicken Diversity Inferred From Mitochondrial DNA.
Godinez CJP, Dadios PJD, Espina DM, Matsunaga M, Nishibori M. Godinez CJP, et al. Front Genet. 2021 Jul 22;12:698401. doi: 10.3389/fgene.2021.698401. eCollection 2021. Front Genet. 2021. PMID: 34367257 Free PMC article.
References
- Dewar RE, Wright HT. 1993. The culture history of Madagascar. J. World Prehist. 7, 417–466. (doi:10.1007/BF00997802) - DOI
- Burney DA, et al. 2004. A chronology for late prehistoric Madagascar. J. Hum. Evol. 47, 25–63. (doi:10.1016/j.jhevol.2004.05.005) - DOI - PubMed
- Beaujard P. 2003. Les arrivées austronésiennes à madagascar: Vagues ou continuum? (partie I). Études Océan Indien. 35–36, 59–147.
- Hurles ME, Sykes BC, Jobling MA, Forster P. 2005. The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am. J. Hum. Genet. 76, 894–901. (doi:10.1086/430051) - DOI - PMC - PubMed
- Tofanelli S, Bertoncini S, Castrì L, Luiselli D, Calafell F, Donati G, Paoli G. 2009. On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 26, 2109–2124. (doi:10.1093/molbev/msp120) - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources