The need for a worldwide consensus for cell line authentication: Experience implementing a mandatory requirement at the International Journal of Cancer - PubMed (original) (raw)
The need for a worldwide consensus for cell line authentication: Experience implementing a mandatory requirement at the International Journal of Cancer
Norbert E Fusenig et al. PLoS Biol. 2017.
Abstract
Cell lines are used in life science research worldwide as biological surrogates. All cell lines are subject to major limitations when used as research tools, including (i) cross-contamination with other cells cultured in the same laboratory environment and (ii) evolution in vitro that renders a given cell line inappropriate as a surrogate for a specific biological hypothesis. There is ample evidence that cross-contamination or phenotypic drift of cells in culture can generate irreproducible or misleading data. A small number of scientific journals-the International Journal of Cancer being at the forefront-and funding agencies have recently moved forward to ask for obligatory cell line authentication data. The history of implementing such rules by the International Journal of Cancer exemplifies the difficulties encountered when installing mandatory quality measures in life sciences.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
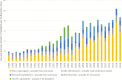
Fig 1. Usage of five misidentified cell lines in the scientific literature as shown by PubMed searches for cell line name and incorrect tissue identity.
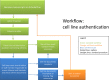
Fig 2. Workflow at the IJC, including checking of cell line documents.
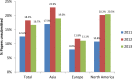
Fig 3. Percentages of papers unsubmitted for cell line issues, by continent and by year.
Generated from S1 Data and S2 Data.
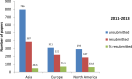
Fig 4. Papers unsubmitted for cell line issues and then resubmitted, by continent in 2011–2013.
Generated from S1 Data.
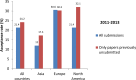
Fig 5. Acceptance rate for all submissions and for papers previously unsubmitted for cell line issues, by continent in 2011–2013.
Generated from S1 Data, S2 Data, and S3 Data.
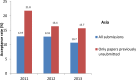
Fig 6. Acceptance rate for all submissions and for papers previously unsubmitted for cell line issues for Asia, by year.
Generated from S1 Data, S2 Data, and S3 Data.
Fig 7. Journal articles using the misidentified cell line HBL-100.
Similar articles
- Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues.
Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kline MC, Kohara A, Los GV, MacLeod RA, Masters JR, Nardone M, Nardone RM, Nims RW, Price PJ, Reid YA, Shewale J, Sykes G, Steuer AF, Storts DR, Thomson J, Taraporewala Z, Alston-Roberts C, Kerrigan L. Barallon R, et al. In Vitro Cell Dev Biol Anim. 2010 Oct;46(9):727-32. doi: 10.1007/s11626-010-9333-z. Epub 2010 Jul 8. In Vitro Cell Dev Biol Anim. 2010. PMID: 20614197 Free PMC article. - Cell line authentication: a necessity for reproducible biomedical research.
Souren NY, Fusenig NE, Heck S, Dirks WG, Capes-Davis A, Bianchini F, Plass C. Souren NY, et al. EMBO J. 2022 Jul 18;41(14):e111307. doi: 10.15252/embj.2022111307. Epub 2022 Jun 27. EMBO J. 2022. PMID: 35758134 Free PMC article. - Knowledge and debate in the American Journal of Clinical Nutrition: new sections, new science, and looking forward and outward.
Duggan CP, Brennan L, Christian P, Fanzo J, Ludwig DS; Editors of the American Journal of Clinical Nutrition. Duggan CP, et al. Am J Clin Nutr. 2020 Jan 1;111(1):1-3. doi: 10.1093/ajcn/nqz267. Am J Clin Nutr. 2020. PMID: 31665209 No abstract available. - The costs of using unauthenticated, over-passaged cell lines: how much more data do we need?
Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. Hughes P, et al. Biotechniques. 2007 Nov;43(5):575, 577-8, 581-2 passim. doi: 10.2144/000112598. Biotechniques. 2007. PMID: 18072586 Review. - Check your cultures! A list of cross-contaminated or misidentified cell lines.
Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Capes-Davis A, et al. Int J Cancer. 2010 Jul 1;127(1):1-8. doi: 10.1002/ijc.25242. Int J Cancer. 2010. PMID: 20143388 Review.
Cited by
- Gynaecological cancers and their cell lines.
Skok K, Gradišnik L, Maver U, Kozar N, Sobočan M, Takač I, Arko D, Kavalar R. Skok K, et al. J Cell Mol Med. 2021 Apr;25(8):3680-3698. doi: 10.1111/jcmm.16397. Epub 2021 Mar 2. J Cell Mol Med. 2021. PMID: 33650759 Free PMC article. Review. - The Imperative Authentication of Cell Lines.
Rojas A. Rojas A. Antimicrob Agents Chemother. 2017 Oct 24;61(11):e01823-17. doi: 10.1128/AAC.01823-17. Print 2017 Nov. Antimicrob Agents Chemother. 2017. PMID: 29066454 Free PMC article. No abstract available. - Feasibility and barriers to rapid establishment of patient-derived primary osteosarcoma cell lines in clinical management.
Chow T, Humble W, Lucarelli E, Onofrillo C, Choong PF, Di Bella C, Duchi S. Chow T, et al. iScience. 2024 Jun 19;27(9):110251. doi: 10.1016/j.isci.2024.110251. eCollection 2024 Sep 20. iScience. 2024. PMID: 39286504 Free PMC article. Review. - Application of RapidHIT™ ID for cell authentication by fast and convenient STR profiling.
Koh UN, Lee JH, Kang HJ, Joo KM, Lee JC, Lim SK. Koh UN, et al. Genes Genomics. 2023 Oct;45(10):1263-1271. doi: 10.1007/s13258-023-01388-4. Epub 2023 May 3. Genes Genomics. 2023. PMID: 37133720 - A comprehensive analysis of e-CAS cell line reveals they are mouse macrophages.
Evans E, Paillot R, López-Álvarez MR. Evans E, et al. Sci Rep. 2018 May 29;8(1):8237. doi: 10.1038/s41598-018-26512-3. Sci Rep. 2018. PMID: 29844485 Free PMC article.
References
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010; 463(7278):184–190. doi: 10.1038/nature08629 - DOI - PMC - PubMed
- Pham K, Delitto D, Knowlton AE, Hartlage ER, Madhavan R, Gonzalo DH, et al. Isolation of pancreatic cancer cells from a patient-derived xenograft model allows for practical expansion and preserved heterogeneity in culture. Am J Pathol. 2016; 186(6):1537–1546. doi: 10.1016/j.ajpath.2016.02.009 - DOI - PMC - PubMed
- Nims RW, Herbstritt CJ. Cell line authentication using isoenzyme analysis. Bio Pharm Int. 2005; 18: 76–82.
- Wrigley JD, McCall EJ, Bannaghan CL, Liggins L, Kendrick C, Griffen A, et al. Cell banking for pharmaceutical research. Drug Discov Today. 2014; 19(10):1518–1529. doi: 10.1016/j.drudis.2014.05.006 - DOI - PubMed
- Characterized Cell Line Core Facility. MD Anderson Cancer Center. https://www.mdanderson.org/education-and-research/resources-for-professi....
MeSH terms
Grants and funding
The author(s) received no specific funding for this work.
LinkOut - more resources
Full Text Sources
Other Literature Sources