Bisulfite-converted duplexes for the strand-specific detection and quantification of rare mutations - PubMed (original) (raw)
Bisulfite-converted duplexes for the strand-specific detection and quantification of rare mutations
Austin K Mattox et al. Proc Natl Acad Sci U S A. 2017.
Abstract
The identification of mutations that are present at low frequencies in clinical samples is an essential component of precision medicine. The development of molecular barcoding for next-generation sequencing has greatly enhanced the sensitivity of detecting such mutations by massively parallel sequencing. However, further improvements in specificity would be useful for a variety of applications. We herein describe a technology (BiSeqS) that can increase the specificity of sequencing by at least two orders of magnitude over and above that achieved with molecular barcoding and can be applied to any massively parallel sequencing instrument. BiSeqS employs bisulfite treatment to distinguish the two strands of molecularly barcoded DNA; its specificity arises from the requirement for the same mutation to be identified in both strands. Because no library preparation is required, the technology permits very efficient use of the template DNA as well as sequence reads, which are nearly all confined to the amplicons of interest. Such efficiency is critical for clinical samples, such as plasma, in which only tiny amounts of DNA are often available. We show here that BiSeqS can be applied to evaluate transversions, as well as small insertions or deletions, and can reliably detect one mutation among >10,000 wild-type molecules.
Keywords: bisulfite sequencing; mutation; next-generation sequencing; polymerase chain reaction; strand-specificity.
Conflict of interest statement
Conflict of interest statement: N.P., K.W.K., and B.V. have no conflicts of interest with respect to the new technology described in this manuscript, as defined by the Johns Hopkins University policy on conflict of interest. N.P., K.W.K., and B.V. are founders of Personal Genome Diagnostics, Inc. and PapGene, Inc. K.W.K. and B.V. are members of the Scientific Advisory Board of Syxmex-Inostics. B.V. is also a member of the Scientific Advisory Boards of Morphotek and Exelixis GP. These companies and others have licensed technologies from Johns Hopkins University; N.P., K.W.K., and B.V. are the inventors of some of these technologies and receive equity or royalties from their licenses. The terms of these arrangements are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Figures
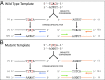
Fig. 1.
Overview of BiSeqS methodology. Bisulfite conversion creates C > T transitions at unique positions in each strand. Amplification of the (+) and (–) strands with primers that are amplicon and strand-specific allows for targeted amplification and addition of molecular barcodes. Analysis of both strands allows for PCR errors generated in the first PCR cycle to be drastically reduced, as it is highly unlikely a complementary mutation will be generated at the same genomic position on both strands. The conversion and amplification of the wild-type sequence is presented in A, and the conversion and amplification of an A > C transversion is presented in B.
Fig. S1.
Detailed schematic of the BiSeqS platform at unmethylated (A) and methylated (B) loci. Unmethylated C is converted to T by bisulfite conversion (step i), and strand-specific PCR-based molecular barcoding adds unique identifiers to the ends of molecules (step ii). Sample barcoding (step iii) amplifies the molecular barcoded DNA, followed by DNA sequencing and analysis (step iv), which allows for the sequences to be aligned to two reference sequences, one for the (+) strand and one for the (–) strand. Universal amplification primers allow for exponential amplification of all barcoded templates, regardless of the UID sequence. The grafting sequences represent the full-length P5 and P7 sequences required for all paired-end reads on Illumina MiSeq platforms.
Fig. S2.
Representative examples of BiSeqS amplicons prepared for eight genomic loci. Differences in primer length often create longer products on one strand, allowing for easy discrimination of equimolar amplification of both strands.
Fig. 2.
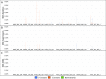
BiSeqS drastically reduces the MAF of single base substitution mutations across amplified loci. (A) MAF of mutations per position across all amplicons. (B) MAF of supermutants per position across all amplicons. (C) MAF of SDMs per position across all amplicons.
Fig. S3.
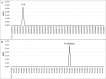
BiSeqS drastically reduces the number of single base substitution mutations. (A) Number of mutations per position across all amplicons. (B) Number of supermutants per position across all amplicons. (C) Number of SDMs per position across all amplicons. Note that the y axis scales in A and C differ by three orders of magnitude.
Fig. S4.
BiSeqS drastically reduces the number of indel mutations across amplified loci. (A) Number of mutations per position across all amplicons. (B) Number of supermutants per position across all amplicons. (C) Number of SDMs per position across all amplicons.
Fig. S5.
BiSeqS drastically reduces the MAF of indel mutations across amplified loci. (A) MAF of mutations per position across all amplicons. (B) MAF of supermutants per position across all amplicons. (C) MAF of SDMs per position across all amplicons.
Fig. 3.
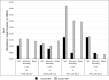
BiSeqS maintains the sensitivity inherent to PCR-based molecular barcoding. Mutant DNA was spiked into normal DNA at a 0.20% or 0.02% target MAF, and the sequencing data were evaluated by standard NGS, molecular barcoding, and BiSeqS.
Fig. S6.
Sensitivity of BiSeqS across all additional amplicons at nominal mutant allele fractions (MAF) of 0.20% and 0.02%. BiSeqS maintains the sensitivity inherent to PCR-based molecular barcoding by detecting mutations at a similar frequency to NGS and molecular barcode-based sequencing.
Fig. S7.
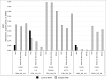
Signal-to-noise plots show that BiSeqS allows for the robust detection of double-strand mutations. (A) A C > A transversion in NRAS at an MAF of 0.20%. (B) A T> deletion in TP53 at an MAF of 0.20%. The actual mutations at the expected positions are detectable in vast excess over background at the other positions using the BiSeqS method.
Similar articles
- Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing.
Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Won HH, et al. J Vis Exp. 2013 Oct 18;(80):e50710. doi: 10.3791/50710. J Vis Exp. 2013. PMID: 24192750 Free PMC article. - Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands.
Cohen JD, Douville C, Dudley JC, Mog BJ, Popoli M, Ptak J, Dobbyn L, Silliman N, Schaefer J, Tie J, Gibbs P, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B. Cohen JD, et al. Nat Biotechnol. 2021 Oct;39(10):1220-1227. doi: 10.1038/s41587-021-00900-z. Epub 2021 May 3. Nat Biotechnol. 2021. PMID: 33941929 Free PMC article. - Fundamentals of pyrosequencing.
Harrington CT, Lin EI, Olson MT, Eshleman JR. Harrington CT, et al. Arch Pathol Lab Med. 2013 Sep;137(9):1296-303. doi: 10.5858/arpa.2012-0463-RA. Arch Pathol Lab Med. 2013. PMID: 23991743 Review. - Clinical mutation assay of tumors: new developments.
Starostik P. Starostik P. Anticancer Drugs. 2017 Jan;28(1):1-10. doi: 10.1097/CAD.0000000000000427. Anticancer Drugs. 2017. PMID: 27575332 Review.
Cited by
- Tumor DNA as a Cancer Biomarker through the Lens of Colorectal Neoplasia.
Cohen JD, Diergaarde B, Papadopoulos N, Kinzler KW, Schoen RE. Cohen JD, et al. Cancer Epidemiol Biomarkers Prev. 2020 Dec;29(12):2441-2453. doi: 10.1158/1055-9965.EPI-20-0549. Epub 2020 Oct 8. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 33033144 Free PMC article. - Next-Generation Genotoxicology: Using Modern Sequencing Technologies to Assess Somatic Mutagenesis and Cancer Risk.
Salk JJ, Kennedy SR. Salk JJ, et al. Environ Mol Mutagen. 2020 Jan;61(1):135-151. doi: 10.1002/em.22342. Epub 2019 Nov 11. Environ Mol Mutagen. 2020. PMID: 31595553 Free PMC article. Review. - The potential of cerebrospinal fluid-based liquid biopsy approaches in CNS tumors.
Mattox AK, Yan H, Bettegowda C. Mattox AK, et al. Neuro Oncol. 2019 Dec 17;21(12):1509-1518. doi: 10.1093/neuonc/noz156. Neuro Oncol. 2019. PMID: 31595305 Free PMC article. Review. - A new approach to decode DNA methylome and genomic variants simultaneously from double strand bisulfite sequencing.
Liang J, Zhang K, Yang J, Li X, Li Q, Wang Y, Cai W, Teng H, Sun Z. Liang J, et al. Brief Bioinform. 2021 Nov 5;22(6):bbab201. doi: 10.1093/bib/bbab201. Brief Bioinform. 2021. PMID: 34058751 Free PMC article. - The Origin of Highly Elevated Cell-Free DNA in Healthy Individuals and Patients with Pancreatic, Colorectal, Lung, or Ovarian Cancer.
Mattox AK, Douville C, Wang Y, Popoli M, Ptak J, Silliman N, Dobbyn L, Schaefer J, Lu S, Pearlman AH, Cohen JD, Tie J, Gibbs P, Lahouel K, Bettegowda C, Hruban RH, Tomasetti C, Jiang P, Chan KCA, Lo YMD, Papadopoulos N, Kinzler KW, Vogelstein B. Mattox AK, et al. Cancer Discov. 2023 Oct 5;13(10):2166-2179. doi: 10.1158/2159-8290.CD-21-1252. Cancer Discov. 2023. PMID: 37565753 Free PMC article.
References
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. - PubMed
- Sidransky D, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. - PubMed
- Sidransky D, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252:706–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA006973/CA/NCI NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- T32 GM007309/GM/NIGMS NIH HHS/United States
- T32 GM007814/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources