The evolutionary history of bears is characterized by gene flow across species - PubMed (original) (raw)
The evolutionary history of bears is characterized by gene flow across species
Vikas Kumar et al. Sci Rep. 2017.
Abstract
Bears are iconic mammals with a complex evolutionary history. Natural bear hybrids and studies of few nuclear genes indicate that gene flow among bears may be more common than expected and not limited to polar and brown bears. Here we present a genome analysis of the bear family with representatives of all living species. Phylogenomic analyses of 869 mega base pairs divided into 18,621 genome fragments yielded a well-resolved coalescent species tree despite signals for extensive gene flow across species. However, genome analyses using different statistical methods show that gene flow is not limited to closely related species pairs. Strong ancestral gene flow between the Asiatic black bear and the ancestor to polar, brown and American black bear explains uncertainties in reconstructing the bear phylogeny. Gene flow across the bear clade may be mediated by intermediate species such as the geographically wide-spread brown bears leading to large amounts of phylogenetic conflict. Genome-scale analyses lead to a more complete understanding of complex evolutionary processes. Evidence for extensive inter-specific gene flow, found also in other animal species, necessitates shifting the attention from speciation processes achieving genome-wide reproductive isolation to the selective processes that maintain species divergence in the face of gene flow.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. Approximate geographic distribution of extant bears according to IUCN data.
Figure has been created using ArcGIS 10 (
http://desktop.arcgis.com/en/arcmap/
) with base map from GADM v 2.0. (
). Species range maps IUCN2015 (
).
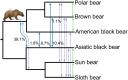
Figure 2. A coalescent species tree and a split network analysis from 18,621 GF ML trees.
(A) In the coalescent species tree all branches receive 100% bootstrap support. The position of root and depicted branch lengths were calculated from coding sequence and 10 Mb of GF data respectively. (B) A split network with a 7% threshold level depicts the complex phylogenetic signal in bear genomes. As expected, the ABC-island brown bear (asterisk) shares alleles with polar bears; among other bears allele sharing is complex. Paintings by Jon Baldur Hlidberg (
).
Figure 3. Phylogenetic relationship among the bears using mtDNA genomes.
A Bayesian tree from 37 complete mt genomes (colored circles) and stars indicate the new mt genomes. The tree is rooted with panda genome (not shown). Supplementary Fig. 12 shows support values for p < 1.0 and accession numbers.
Figure 4. Graphical summary of gene flow analyses using D and D FOIL statistics on a cladogram.
D FOIL analyses estimated the percentage of GFs rejecting the species tree and indicating gene flow. Blue arrows show values >1%, and dashed lavender for <0.1% (Table 1). These percentages do not indicate the amount of introgressed DNA, which can be a fraction of the GF sequence. Green arrows depict significant _D_-statistics data for gene flow signal. Some gene flow cannot have occurred directly between species, because the species exist in different habitats, but may be remnants of ancestral gene flow or gene flow through a vector species.
Figure 5. Phylogenomic estimates of divergence times.
The scale bar shows divergence times in million years and 95% confidence intervals for divergence times are shown as shadings (Supplementary Table 7). The tree is rooted with the panda genome (not shown).
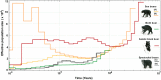
Figure 6. Historical effective population sizes (N e) using the pairwise Markovian coalescent (PSMC) analyses for the newly sequenced bear genomes.
X-axis:time, y-axis:effective population size (N e). The two sun bears have radically different, non-overlapping population histories (Supplementary Fig. 21). The Asiatic black bear had a constant large Ne since 500 ka similar to that of the brown bear and consistent with a wide geographic distribution and high heterozygosity (Supplementary Fig. 4).
Similar articles
- Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears.
Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B. Cahill JA, et al. Mol Ecol. 2015 Mar;24(6):1205-17. doi: 10.1111/mec.13038. Epub 2015 Feb 5. Mol Ecol. 2015. PMID: 25490862 Free PMC article. - Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage.
Hailer F, Kutschera VE, Hallström BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. Hailer F, et al. Science. 2012 Apr 20;336(6079):344-7. doi: 10.1126/science.1216424. Science. 2012. PMID: 22517859 - Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution.
Cahill JA, Green RE, Fulton TL, Stiller M, Jay F, Ovsyanikov N, Salamzade R, St John J, Stirling I, Slatkin M, Shapiro B. Cahill JA, et al. PLoS Genet. 2013;9(3):e1003345. doi: 10.1371/journal.pgen.1003345. Epub 2013 Mar 14. PLoS Genet. 2013. PMID: 23516372 Free PMC article. - The Net of Life, a short story: Intricate patterns of gene flows across hundreds of extant genomes, all the way to LUCA.
Ouzounis CA. Ouzounis CA. Biosystems. 2024 May;239:105199. doi: 10.1016/j.biosystems.2024.105199. Epub 2024 Apr 17. Biosystems. 2024. PMID: 38641198 Review. - Sequencing and assembling bear genomes: the bare necessities.
Willey C, Korstanje R. Willey C, et al. Front Zool. 2022 Nov 30;19(1):30. doi: 10.1186/s12983-022-00475-8. Front Zool. 2022. PMID: 36451195 Free PMC article. Review.
Cited by
- The role of introgression and ecotypic parallelism in delineating intraspecific conservation units.
Taylor RS, Manseau M, Horn RL, Keobouasone S, Golding GB, Wilson PJ. Taylor RS, et al. Mol Ecol. 2020 Aug;29(15):2793-2809. doi: 10.1111/mec.15522. Epub 2020 Jul 11. Mol Ecol. 2020. PMID: 32567754 Free PMC article. - A most aggressive bear: Safari videos document sloth bear defense against tiger predation.
Sharp TR, Garshelis DL, Larson W. Sharp TR, et al. Ecol Evol. 2024 Jul 11;14(7):e11524. doi: 10.1002/ece3.11524. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39005887 Free PMC article. - Speciation Generates Mosaic Genomes in Kangaroos.
Nilsson MA, Zheng Y, Kumar V, Phillips MJ, Janke A. Nilsson MA, et al. Genome Biol Evol. 2018 Jan 1;10(1):33-44. doi: 10.1093/gbe/evx245. Genome Biol Evol. 2018. PMID: 29182740 Free PMC article. - Genome-wide analysis reveals signatures of complex introgressive gene flow in macaques (genus Macaca).
Song Y, Jiang C, Li KH, Li J, Qiu H, Price M, Fan ZX, Li J. Song Y, et al. Zool Res. 2021 Jul 18;42(4):433-449. doi: 10.24272/j.issn.2095-8137.2021.038. Zool Res. 2021. PMID: 34114757 Free PMC article. - The Syngameon Enigma.
Buck R, Flores-Rentería L. Buck R, et al. Plants (Basel). 2022 Mar 28;11(7):895. doi: 10.3390/plants11070895. Plants (Basel). 2022. PMID: 35406874 Free PMC article. Review.
References
- Wagner J. Pliocene to early Middle Pleistocene ursine bears in Europe: a taxonomic overview. J. Natl. Mus. Prague Nat. Hist. Ser. 179, 197–215 (2010).
- Coyne J. A. & Orr H. A. Speciation. 37, (Sunderland, MA: Sinauer Associates, 2004).
- Gray A. Mammalian hybrids. A check-list with bibliography. (Commonwealth Agricultural Bureaux, 1972).
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources