Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23 - PubMed (original) (raw)
Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23
See-Yeun Ting et al. Elife. 2017.
Abstract
Proteins destined for the mitochondrial matrix are targeted to the inner membrane Tim17/23 translocon by their presequences. Inward movement is driven by the matrix-localized, Hsp70-based motor. The scaffold Tim44, interacting with the matrix face of the translocon, recruits other motor subunits and binds incoming presequence. The basis of these interactions and their functional relationships remains unclear. Using site-specific in vivo crosslinking and genetic approaches in Saccharomyces cerevisiae, we found that both domains of Tim44 interact with the major matrix-exposed loop of Tim23, with the C-terminal domain (CTD) binding Tim17 as well. Results of in vitro experiments showed that the N-terminal domain (NTD) is intrinsically disordered and binds presequence near a region important for interaction with Hsp70 and Tim23. Our data suggest a model in which the CTD serves primarily to anchor Tim44 to the translocon, whereas the NTD is a dynamic arm, interacting with multiple components to drive efficient translocation.
Keywords: Hsp70; S. cerevisiae; biochemistry; cell biology; mitochondria; protein translocation.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
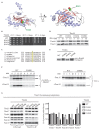
Figure 1.. Residue D345 in the CTD of Tim44 is important for cell viability and association of the import motor with the translocon.
(A) Structure of the CTD of Tim44; PDB entry 2FXT (Josyula et al., 2006). α-helices and β-strands are labeled. Residue D345 is indicated and shown in sphere representation. Right view of the structure, rotated 90° along the horizontal axis from the orientation on the left. (B) Growth phenotype. Ten-fold serial dilutions of _tim44_-Δ cells expressing the indicated Tim44 variants were plated on minimal media and incubated at the indicated temperatures and times. (C) Sequence alignment of Tim44 from indicated eukaryotic species was done using Clustal Omega; residue D345 is indicated (*). (D) 5 and 15 μg of mitochondria isolated from Tim44 mutants were analyzed by SDS–PAGE, followed by immunoblotting against Tim23-and Tim44-specific antibodies. (E) Precursor accumulation. Whole cell lysates of the indicated TIM44 mutants were prepared from cells grown at 30°C and shifted to 37°C for 6 hr. Proteins separated by SDS–PAGE were subjected to immunoblot analysis using antibodies specifiic to Hsp60. The Hsp60 precursor form (pre-Hsp60) and mature form (mHsp60) are indicated. (F) 35S-labeled precursors of cytochrome b2-(167)Δ19-DHFR and cytochrome c1 were imported into wild-type (WT) and tim44D345K mutant mitochondria at 25°C. Where indicated, mitochondria were subsequently treated with proteinase K (Prot. K). Samples were analyzed by SDS-PAGE and autoradiography. p, precursor; I, intermediate; m, mature. (G) Co-immunoprecipitation with Tim23. Mitochondria were solubilized by treatment with digitonin. Solubilized material was subjected to immunoprecipitation using Tim23-specific antibodies crosslinked to protein A beads. Precipitates were analyzed by SDS-PAGE and immunoblotting using antibodies specific for the indicated proteins. 10% of input was used as a loading control. Signals were quantified by Image J software (RRID:
SCR_003070
) and plotted as percentages of the wt control reaction value. Data represent the mean ± standard deviation, with n = 3. The data of the figure can also be seen in Figure 1—source data 1. DOI:
http://dx.doi.org/10.7554/eLife.23609.002
Figure 2.. Importance of conserved residues near D345.
(A) Structure of the CTD of Tim44; PDB entry 2FXT (Josyula et al., 2006). The five nearby charged residues analyzed (E295, E303, R342, E350, and R370) are indicated. (B) Growth phenotype. Ten-fold serial dilutions of _tim44_-Δ cells expressing the indicated Tim44 variants were plated on minimal media and incubated at the indicated temperatures and times. (C) 5 and 15 μg of mitochondria isolated from Tim44 mutants were analyzed by SDS–PAGE, followed by immunoblotting against antibodies to Tim23 and Tim44. (D) Precursor accumulation. Whole cell lysates from indicated cells grown at 30°C and shifted to 37°C for 6 hr (left). tim44D345A/E303A was grown at 23°C and then shifted to 37°C for 6 hr, because it does not grow at 30°C (right). Proteins separated by SDS–PAGE were subjected to immunoblot analysis using antibodies specific to Hsp60. The Hsp60 precursor form (pre-Hsp60) and mature form (mHsp60) are indicated. (E) Co-immunoprecipitation with Tim23. Mitochondria were solubilized by treatment with digitonin. Solubilized material was subjected to immunoprecipitation using Tim23-specific antibodies crosslinked to protein A beads. Precipitates were analyzed by SDS-PAGE and immunoblotting using antibodies specific for the indicated proteins. 10% of input for immunoprecipitation was used as a loading control. Signals were quantified by Image J (RRID:
SCR_003070
) software and plotted as percentages of the wild-type control reaction value. Data represent the mean ± standard deviation, with n = 3. The data of the figure can also be seen in Figure 2—source data 1. (F) 35S-labeled precursors of cytochrome b2-(167)Δ19-DHFR and cytochrome c1 were imported into wild-type (WT) and tim44E303A/D345A mutant mitochondria at 25°C. Where indicated, mitochondria were subsequently treated with proteinase K (Prot. K). Samples were analyzed by SDS-PAGE and autoradiography. p, precursor; I, intermediate; m, mature. (G) Growth phenotype. Ten-fold serial dilutions of _tim44_-Δ cells expressing the indicated Tim44 variants were plated on minimal media and incubated at the indicated temperatures and times. DOI:
http://dx.doi.org/10.7554/eLife.23609.004
Figure 2—figure supplement 1.. Sequence alignment of Tim44 from indicated eukaryotic species was done using Clustal Omega.
Residue E295, E303, R342, D345, E350, R370 are labeled in red, with the endpoint residue numbers indicated. DOI:
http://dx.doi.org/10.7554/eLife.23609.006
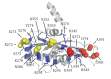
Figure 3.. Site-specific crosslinking of Tim44 to Tim23 or Tim17.
(A,B,C). Yeast strains expressing variants of Tim44 with Bpa incorporated at the indicated positions. After UV irradiation (+), or as a control no irradiation (-), Tim44 was affinity purified via its N-terminal His6 tag, followed by SDS-PAGE and immunoblotting with the indicated antibodies. Migrations of size standard, in kDa, are indicted (left); Tim44 (44) and Tim23 (23) or Tim17 (17) reactive bands are indicated. The asterisks indicate non-specific cross-reactive bands. (D) The residues whose crosslinking was detected are shown in sphere representation (Red, Tim44-Tim23 crosslinks; yellow, Tim44-Tim17 crosslinks). DOI:
http://dx.doi.org/10.7554/eLife.23609.007
Figure 3—figure supplement 1.. Summary of Tim44 CTD in vivo crosslinking.
The residues whose crosslinking was detected are shown in sphere representation. Red, Tim44-Tim23 crosslinks (D345Bpa, I374Bpa, I385Bpa, A391Bpa); yellow, Tim44-Tim17 crosslinks (S271Bpa, R272Bpa, N286Bpa, E350Bpa). The residues whose crosslinking was not significant are shown in stick representation (blue): S270, Y274, S275, K278, E387, R291, E295, E303, L344, R342, R347, V352, K355, R370, Q372. DOI:
http://dx.doi.org/10.7554/eLife.23609.008
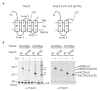
Figure 4.. Both domains of Tim44 crosslink to loop 1 of Tim23.
(A) Diagram of Tim23 (left) and Tim23Δ176-222 (right). Loops, transmembrane domains (TM), and residue 175 are indicated. (B) Yeast strains expressing Tim23 or Tim23Δ176-222 and Tim44 with Bpa incorporated at position 165 in the NTD or 391 in the CTD were subjected to UV irradiation (+), or as a control not exposed (-). Tim44 was then affinity purified via its N-terminal His6 tag, followed by SDS-PAGE and immunoblotting with the indicated antibodies. Migration of size standards, in kDa, are indicted (left); Tim44 (44NTD or 44CTD) and Tim23 (23 or 23ΔCTD) reactive bands are indicated. The asterisk indicates non-specific cross-reactive bands. DOI:
http://dx.doi.org/10.7554/eLife.23609.009
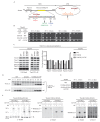
Figure 5.. Tim44 NTD alteration suppresses D345K CTD defects.
(A) Diagram of Tim44 showing functional regions and interaction partners of the NTD. Residues from 156 to 184 are indicated in yellow. Residues crosslinked to Tim23 in our previous report (Ting et al., 2014) are in red; residue G173, the suppressor reported here, and R180, the previously isolated ts mutant (Schiller et al., 2008), which affects Tim23 and mtHsp70 association, are in green and blue, respectively. The N terminal segment of Tim44 (residues 51 to 82) implicated in interaction with Pam16 is indicated (Schilke et al., 2012). The mutant D345A/E350A and residues D165Bpa, S271Bpa, A391Bpa used for crosslinking in combination the G173V suppressing substitution are indicated in orange and dark red, respectively. (B) Growth phenotype. Ten-fold serial dilutions of _tim44_-Δ cells expressing Tim44 variants were plated on minimal media and incubated at the indicated temperatures and times. For each temperature, all strains were plated on the same plate; strains not relevant to this report were cropped out (indicated by white space). (C) Co-immunoprecipitation with Tim23. Mitochondria were solubilized by treatment with digitonin. Solubilized material was subjected to immunoprecipitation using Tim23-specific antibodies crosslinked to protein A beads. Precipitates were analyzed by SDS-PAGE and immunoblotting using antibodies specific for the indicated proteins. 10% of input for co-immunoprecipitation was used as a loading control. Signals were quantified by Image J software (RRID:
SCR_003070
) and plotted as percentages of the wt control reaction value. Data represent the mean ± standard deviation, with n = 3. The data of the figure can also be seen in Figure 5—source data 1. (D) 35S-labeled precursor of cytochrome b2-(167)Δ19-DHFR was imported into wild-type (WT), tim44D345AK, and tim44D345AK/G173V mitochondria at 25°C. Where indicated, mitochondria were subsequently treated with proteinase K (Prot. K). Samples were analyzed by SDS-PAGE and autoradiography. p, precursor; I, intermediate; m, mature. (E) Growth phenotype. Ten-fold serial dilutions of _tim44_-Δ cells expressing the indicated Tim44 variants were plated on minimal media and incubated at the temperatures and for the times indicated. (F) Yeast strains expressing tim44D345A/E350A with Bpa incorporated at position D165, S271 or A391 were subjected to UV irradiation (+), or as a control not exposed (-). Tim44 was then affinity purified via its N-terminal His6 tag, followed by SDS-PAGE and immunoblotting with the indicated antibodies. Migration of size standards, in kDa, are indicated (left); Tim44 (44NTD or 44CTD), Tim23 (23), and Tim17 (17) reactive bands are indicated. The asterisk indicates non-specific cross-reactive bands. DOI:
http://dx.doi.org/10.7554/eLife.23609.010
Figure 6.. Interaction of the NTD of Tim44 with presequence.
(A) Diagram of Tim44 showing different constructs along with a summary of crosslinking efficiency to the presequence of Hsp60 (pHsp60). Endpoint residue numbers are indicated. (B,D,E left) Crosslinking was performed by incubating biotin-labeled pHsp60 with indicated Tim44 proteins in the absence (-) or presence (+) of disuccinimidyl glutarate (DSG), followed by SDS-PAGE and immunoblotting using antibodies specific for biotin (streptavidin-HRP). (B,D,E right) Equal molar amounts of indicated Tim44 proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. (B) WT, full length Tim44 (residues 43–431); NTD (residues 43–209); CTD (residues 210–431). (C) Crosslinking of Tim4443-209 with indicated increasing concentration of pHsp60. The asterisk indicates higher crosslinking products of the Tim4443-209 -pHsp60 complex. (D) Crosslinking of NTD and indicated C-terminal truncations. (E) Crosslinking of NTD and indicated internal deletions. DOI:
http://dx.doi.org/10.7554/eLife.23609.012
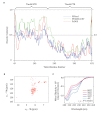
Figure 7.. Evidence that the NTD of Tim44 is intrinsically disordered.
(A) Prediction of disorder by indicated disorder predictors. The NTD and CTD of Tim44 are indicated at the top of the graph. Residues with a disorder tendency exceeding 0.5 were considered to be disordered. The data of the figure can also be seen in Figure 7—source data 1. (B) 2D 1H,15N-Heteronuclear Single-Quantum Correlation (HSQC) NMR spectra of Tim4443-209. As described in the text, the low dispersion of 1HN chemical shifts is indicative of an unfolded protein in solution. (C) Far-UV CD spectra of Tim4443-209 as a function of temperature, recorded over the temperature range from 4°C to 80°C. After measuring the sample at 80°C, it was cooled down to 4°C to test the reversibility (4°C return). Smoothing spline model was used to fit the raw CD data. The data of the figure can also be seen in Figure 7—source data 2. DOI:
http://dx.doi.org/10.7554/eLife.23609.013
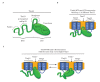
Figure 8.. Diagrams of Tim44 interaction with the translocon.
(A) Diagram of Tim44 (green) with the regions known to be important for interaction with other motor or translocon components indicated by dotted circles. The NTD is drawn to indicate its naturally disordered state. (B) Model of Tim44 interaction with loop 1 of two different Tim23 molecules (blue). Transmembrane domains 1 and 2 of Tim23 are indicated. Tim17 is indicated in orange. (C) Model of Tim44 NTD and CTD interacting with loop 1 of one Tim23 molecule, either the NTD (left) or CTD (right). DOI:
http://dx.doi.org/10.7554/eLife.23609.016
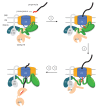
Figure 9.. Model of Tim44 action during protein translocation.
As in Figure 8C, one Tim44 is drawn as interacting with one Tim23 molecule. Pam16 interacts with the extreme N-terminus of the Tim44 NTD, and also interacts with the J-protein co-chaperone Pam18 (‘J’ indicates its J domain). Steps 1–4 are discussed in the discussion section of the text. Tim44 (green), with CTD (oval) and NTD (line); Tim 23 (blue); Tim17 (orange); Presequence (red line); remainder of translocating polypeptide chain (black line). DOI:
http://dx.doi.org/10.7554/eLife.23609.017
Similar articles
- Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor.
Ting SY, Schilke BA, Hayashi M, Craig EA. Ting SY, et al. J Biol Chem. 2014 Oct 10;289(41):28689-96. doi: 10.1074/jbc.M114.588152. Epub 2014 Aug 25. J Biol Chem. 2014. PMID: 25157107 Free PMC article. - Role of Tim17 in coupling the import motor to the translocation channel of the mitochondrial presequence translocase.
Demishtein-Zohary K, Günsel U, Marom M, Banerjee R, Neupert W, Azem A, Mokranjac D. Demishtein-Zohary K, et al. Elife. 2017 Feb 6;6:e22696. doi: 10.7554/eLife.22696. Elife. 2017. PMID: 28165323 Free PMC article. - Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase.
Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M. Hutu DP, et al. Mol Biol Cell. 2008 Jun;19(6):2642-9. doi: 10.1091/mbc.e07-12-1226. Epub 2008 Apr 9. Mol Biol Cell. 2008. PMID: 18400944 Free PMC article. - On the mechanism of preprotein import by the mitochondrial presequence translocase.
van der Laan M, Hutu DP, Rehling P. van der Laan M, et al. Biochim Biophys Acta. 2010 Jun;1803(6):732-9. doi: 10.1016/j.bbamcr.2010.01.013. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20100523 Review. - The preprotein translocase of the mitochondrial inner membrane: function and evolution.
Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. Rassow J, et al. J Mol Biol. 1999 Feb 12;286(1):105-20. doi: 10.1006/jmbi.1998.2455. J Mol Biol. 1999. PMID: 9931253 Review.
Cited by
- Protein import motor complex reacts to mitochondrial misfolding by reducing protein import and activating mitophagy.
Michaelis JB, Brunstein ME, Bozkurt S, Alves L, Wegner M, Kaulich M, Pohl C, Münch C. Michaelis JB, et al. Nat Commun. 2022 Sep 2;13(1):5164. doi: 10.1038/s41467-022-32564-x. Nat Commun. 2022. PMID: 36056001 Free PMC article. - NAC and Zuotin/Hsp70 chaperone systems coexist at the ribosome tunnel exit in vivo.
Ziegelhoffer T, Verma AK, Delewski W, Schilke BA, Hill PM, Pitek M, Marszalek J, Craig EA. Ziegelhoffer T, et al. Nucleic Acids Res. 2024 Apr 12;52(6):3346-3357. doi: 10.1093/nar/gkae005. Nucleic Acids Res. 2024. PMID: 38224454 Free PMC article. - Impact of Mitophagy and Mitochondrial Unfolded Protein Response as New Adaptive Mechanisms Underlying Old Pathologies: Sarcopenia and Non-Alcoholic Fatty Liver Disease.
Urbina-Varela R, Castillo N, Videla LA, Del Campo A. Urbina-Varela R, et al. Int J Mol Sci. 2020 Oct 18;21(20):7704. doi: 10.3390/ijms21207704. Int J Mol Sci. 2020. PMID: 33081022 Free PMC article. Review. - The ADP/ATP translocase drives mitophagy independent of nucleotide exchange.
Hoshino A, Wang WJ, Wada S, McDermott-Roe C, Evans CS, Gosis B, Morley MP, Rathi KS, Li J, Li K, Yang S, McManus MJ, Bowman C, Potluri P, Levin M, Damrauer S, Wallace DC, Holzbaur ELF, Arany Z. Hoshino A, et al. Nature. 2019 Nov;575(7782):375-379. doi: 10.1038/s41586-019-1667-4. Epub 2019 Oct 16. Nature. 2019. PMID: 31618756 Free PMC article. - Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences.
Backes S, Hess S, Boos F, Woellhaf MW, Gödel S, Jung M, Mühlhaus T, Herrmann JM. Backes S, et al. J Cell Biol. 2018 Apr 2;217(4):1369-1382. doi: 10.1083/jcb.201708044. Epub 2018 Jan 30. J Cell Biol. 2018. PMID: 29382700 Free PMC article.
References
- Blom J, Kübrich M, Rassow J, Voos W, Dekker PJ, Maarse AC, Meijer M, Pfanner N. The essential yeast protein MIM44 (encoded by MPI1) is involved in an early step of preprotein translocation across the mitochondrial inner membrane. Molecular and Cellular Biology. 1993;13:7364–7371. doi: 10.1128/MCB.13.12.7364. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- P41 RR002301/RR/NCRR NIH HHS/United States
- S10 RR013790/RR/NCRR NIH HHS/United States
- R01 GM027870/GM/NIGMS NIH HHS/United States
- S10 RR025062/RR/NCRR NIH HHS/United States
- S10 RR029220/RR/NCRR NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- P41 GM103399/GM/NIGMS NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
- S10 RR023438/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases