Comparative interactomics provides evidence for functional specialization of the nuclear pore complex - PubMed (original) (raw)
Comparative interactomics provides evidence for functional specialization of the nuclear pore complex
Samson O Obado et al. Nucleus. 2017.
Abstract
The core architecture of the eukaryotic cell was established well over one billion years ago, and is largely retained in all extant lineages. However, eukaryotic cells also possess lineage-specific features, frequently keyed to specific functional requirements. One quintessential core eukaryotic structure is the nuclear pore complex (NPC), responsible for regulating exchange of macromolecules between the nucleus and cytoplasm as well as acting as a nuclear organizational hub. NPC architecture has been best documented in one eukaryotic supergroup, the Opisthokonts (e.g. Saccharomyces cerevisiae and Homo sapiens), which although compositionally similar, have significant variations in certain NPC subcomplex structures. The variation of NPC structure across other taxa in the eukaryotic kingdom however, remains poorly understood. We explored trypanosomes, highly divergent organisms, and mapped and assigned their NPC proteins to specific substructures to reveal their NPC architecture. We showed that the NPC central structural scaffold is conserved, likely across all eukaryotes, but more peripheral elements can exhibit very significant lineage-specific losses, duplications or other alterations in their components. Amazingly, trypanosomes lack the major components of the mRNA export platform that are asymmetrically localized within yeast and vertebrate NPCs. Concomitant with this, the trypanosome NPC is ALMOST completely symmetric with the nuclear basket being the only major source of asymmetry. We suggest these features point toward a stepwise evolution of the NPC in which a coating scaffold first stabilized the pore after which selective gating emerged and expanded, leading to the addition of peripheral remodeling machineries on the nucleoplasmic and cytoplasmic sides of the pore.
Keywords: Trypanosoma brucei; eukaryogenesis; mRNA export; molecular evolution; nuclear pore complex.
Figures
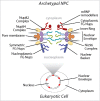
Figure 1.
Schematic of opisthokont NPCs. A schematic of textbook NPCs highlighting each distinct nuclear pore subcomplex. The approximate location messenger ribonucleprotein (mRNP) remodeling factors required for mRNA export out of the NPC is also shown.
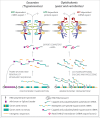
Figure 2.
(A) comparison between Excavate and Opisthokont NPCs and transport through them. Trypanosomes (Excavates) have a symmetric NPC with the exception of the nuclear basket unlike in opisthokonts (yeast and humans). Furthermore, trypanosomes lack the cytoplasmic mRNA platform including mRNP (ribonucleoproteins) remodeling factors such as the ATP-dependent DEAD box helicase Dbp5 and Gle1 that are crucial for mRNA export in opisthokonts. Instead, mRNA export in trypanosomes appears to rely on the RanGDP/GTP gradient similar to protein export, as opposed to ATP in opisthokonts. This may be related to the unusual mechanisms trypanosomes use for controlling gene expression. Trypanosome protein-coding genes are intronless and each gene lacks an individual polymerase II promoter. Thus, trypanosome genes (green boxes) are transcribed into long multigene (polycistronic) transcripts that are resolved into single mRNAs by _trans_-splicing of a mini exon, also known as the spliced leader sequence (purple box) at the 5′ end of each gene, splicing out of intergenic (orange lines) sequences and polyadenylated (AAA). Processed and export competent mRNA are exported through the NPC by the Mex67/Mtr2 heterodimer (pink ovals). Protein coding genes in opisthokonts (and most other eukaryotes) have introns (orange boxes) and individual promoters (flags) and are transcribed as singly (monocistronic). Transcribed mRNAs are co-transcriptionally 5′-capped (gray circles), and introns spliced (orange lines) before export by Mex67/Mtr2 in conjunction with the actions of the ATP dependent helicase Dbp5.
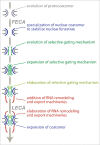
Figure 3.
Evolution of the NPC. We propose a model in which the NPC evolved gating functions in a stepwise manner starting with a simple coat that acquired complexity through a series of duplications as observed in endomembrane trafficking. This then led to the evolution of FG-Nups and then further diversification into the current metazoan type NPC with a nuclear basket and cytoplasmic filaments.
Comment on
- Extra view to: Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol. 2016; 14(2):e1002365; PMID:; https://doi.org/10.1371/journal.pbio.1002365.
Similar articles
- A lineage-specific protein network at the trypanosome nuclear envelope.
Butterfield ER, Obado SO, Scutts SR, Zhang W, Chait BT, Rout MP, Field MC. Butterfield ER, et al. Nucleus. 2024 Dec;15(1):2310452. doi: 10.1080/19491034.2024.2310452. Epub 2024 Apr 11. Nucleus. 2024. PMID: 38605598 Free PMC article. - Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex.
Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Obado SO, et al. PLoS Biol. 2016 Feb 18;14(2):e1002365. doi: 10.1371/journal.pbio.1002365. eCollection 2016 Feb. PLoS Biol. 2016. PMID: 26891179 Free PMC article. - Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor.
DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. DeGrasse JA, et al. Mol Cell Proteomics. 2009 Sep;8(9):2119-30. doi: 10.1074/mcp.M900038-MCP200. Epub 2009 Jun 13. Mol Cell Proteomics. 2009. PMID: 19525551 Free PMC article. - Enriching the pore: splendid complexity from humble origins.
Field MC, Koreny L, Rout MP. Field MC, et al. Traffic. 2014 Feb;15(2):141-56. doi: 10.1111/tra.12141. Epub 2014 Jan 8. Traffic. 2014. PMID: 24279500 Free PMC article. Review. - Evolutionary, structural and functional insights in nuclear organisation and nucleocytoplasmic transport in trypanosomes.
Padilla-Mejia NE, Field MC. Padilla-Mejia NE, et al. FEBS Lett. 2023 Oct;597(20):2501-2518. doi: 10.1002/1873-3468.14747. Epub 2023 Oct 15. FEBS Lett. 2023. PMID: 37789516 Free PMC article. Review.
Cited by
- The Trypanosoma brucei RNA-binding protein DRBD18 ensures correct mRNA trans splicing and polyadenylation patterns.
Bishola Tshitenge T, Clayton C. Bishola Tshitenge T, et al. RNA. 2022 Sep;28(9):1239-1262. doi: 10.1261/rna.079258.122. Epub 2022 Jul 6. RNA. 2022. PMID: 35793904 Free PMC article. - Pore timing: the evolutionary origins of the nucleus and nuclear pore complex.
Field MC, Rout MP. Field MC, et al. F1000Res. 2019 Apr 3;8:F1000 Faculty Rev-369. doi: 10.12688/f1000research.16402.1. eCollection 2019. F1000Res. 2019. PMID: 31001417 Free PMC article. Review. - Nuclear mRNA maturation and mRNA export control: from trypanosomes to opisthokonts.
Kramer S. Kramer S. Parasitology. 2021 Sep;148(10):1196-1218. doi: 10.1017/S0031182021000068. Epub 2021 Jan 19. Parasitology. 2021. PMID: 33461637 Free PMC article. Review. - Trypanosomes can initiate nuclear export co-transcriptionally.
Goos C, Dejung M, Wehman AM, M-Natus E, Schmidt J, Sunter J, Engstler M, Butter F, Kramer S. Goos C, et al. Nucleic Acids Res. 2019 Jan 10;47(1):266-282. doi: 10.1093/nar/gky1136. Nucleic Acids Res. 2019. PMID: 30418648 Free PMC article. - A lineage-specific protein network at the trypanosome nuclear envelope.
Butterfield ER, Obado SO, Scutts SR, Zhang W, Chait BT, Rout MP, Field MC. Butterfield ER, et al. Nucleus. 2024 Dec;15(1):2310452. doi: 10.1080/19491034.2024.2310452. Epub 2024 Apr 11. Nucleus. 2024. PMID: 38605598 Free PMC article.
References
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biology letters 2010; 6:342-5; PMID:20031978; https://doi.org/10.1098/rsbl.2009.0948 - DOI - PMC - PubMed
- Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al.. The revised classification of eukaryotes. J Eukaryot Microbiol 2012; 59:429-93; PMID:23020233; https://doi.org/10.1111/j.1550-7408.2012.00644.x - DOI - PMC - PubMed
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158:915-27; PMID:12196509; https://doi.org/10.1083/jcb.200206106 - DOI - PMC - PubMed
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics : MCP 2009; 8:2119-30; https://doi.org/10.1074/mcp.M900038-MCP200 - DOI - PMC - PubMed
- Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol 2016; 14:e1002365; PMID:26891179; https://doi.org/10.1371/journal.pbio.1002365 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 203134/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/N010558/1/MRC_/Medical Research Council/United Kingdom
- P41 GM109824/GM/NIGMS NIH HHS/United States
- R01 GM112108/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources