Protein composition of the hepatitis A virus quasi-envelope - PubMed (original) (raw)
Protein composition of the hepatitis A virus quasi-envelope
Kevin L McKnight et al. Proc Natl Acad Sci U S A. 2017.
Abstract
The Picornaviridae are a diverse family of RNA viruses including many pathogens of medical and veterinary importance. Classically considered "nonenveloped," recent studies show that some picornaviruses, notably hepatitis A virus (HAV; genus Hepatovirus) and some members of the Enterovirus genus, are released from cells nonlytically in membranous vesicles. To better understand the biogenesis of quasi-enveloped HAV (eHAV) virions, we conducted a quantitative proteomics analysis of eHAV purified from cell-culture supernatant fluids by isopycnic ultracentrifugation. Amino acid-coded mass tagging (AACT) with stable isotopes followed by tandem mass spectrometry sequencing and AACT quantitation of peptides provided unambiguous identification of proteins associated with eHAV versus unrelated extracellular vesicles with similar buoyant density. Multiple peptides were identified from HAV capsid proteins (53.7% coverage), but none from nonstructural proteins, indicating capsids are packaged as cargo into eHAV vesicles via a highly specific sorting process. Other eHAV-associated proteins (n = 105) were significantly enriched for components of the endolysosomal system (>60%, P < 0.001) and included many common exosome-associated proteins such as the tetraspanin CD9 and dipeptidyl peptidase 4 (DPP4) along with multiple endosomal sorting complex required for transport III (ESCRT-III)-associated proteins. Immunoprecipitation confirmed that DPP4 is displayed on the surface of eHAV produced in cell culture or present in sera from humans with acute hepatitis A. No LC3-related peptides were identified by mass spectrometry. RNAi depletion studies confirmed that ESCRT-III proteins, particularly CHMP2A, function in eHAV biogenesis. In addition to identifying surface markers of eHAV vesicles, the results support an exosome-like mechanism of eHAV egress involving endosomal budding of HAV capsids into multivesicular bodies.
Keywords: ESCRT; exosome; extracellular vesicle; multivesicular body; picornavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
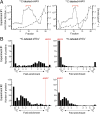
Stable isotope labeling of eHAV. (A) Distribution of HAV RNA (log10 scale) in fractions collected from iodixanol gradients loaded with eHAV concentrated from supernatant fluids of infected Huh-7 cell cultures maintained in media containing H [13C] (Left) or L [12C] (Right) amino acids in experiment 1 and centrifuged to equilibrium. The red brackets indicate fractions that were pooled with fractions of similar density from gradients loaded with supernatant fluids from noninfected cells maintained in the opposing media and subjected to mass spectrometry. ○, HAV RNA; GEs (genome equivalents) per 20 µL; ♦, density (g/cm3). (B) Distribution of isotopic enrichment in peptides identified in a search against the human UniProt database in the four independently 13C-labeled (Left) and 12C-labeled (Right) eHAV samples submitted for mass spectrometry in experiments 1 (Top) and 2 (Bottom). In experiment 1, infected and uninfected cell-culture harvests were mixed following density gradient fractionation, whereas these samples were mixed before fractionation in experiment 2. The fold enrichment of HAV-encoded peptides is noted in each panel.
Fig. S1.
HAV peptide sequence coverage. The polyprotein sequence of HM175/p16 virus (GenBank accession no. KP879217.1) is shown with capsid proteins (VP4 to pX) (Top) and nonstructural proteins (2B to 3Dpol) (Bottom). Amino acid residues shaded in green represent those in peptides identified by mass spectrometry in any of the four independently labeled eHAV samples. Residues shaded in magenta represent individual peptides spanning the VP4–VP2 maturation cleavage and VP1–pX cleavage sites. Overall peptide coverage was 53.7% for capsid (structural) proteins and 0% for nonstructural proteins.
Fig. S2.
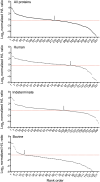
Distribution of automated MaxQuant normalized H/L ratios for human and bovine protein groups identified by quantifiable peptides (pooled results from four virus samples in two independent experiments). From the top down: all proteins (n = 460), uniquely human (n = 154), indeterminate species of origin (n = 172), and uniquely bovine (n = 134). The dashed horizontal line in each panel represents the median H/L ratio for all proteins. Arrows indicate the point at which the distribution crosses the median H/L ratio value of all proteins.
Fig. 2.
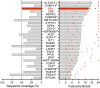
Sequence coverage and geometric mean fold isotope enrichment of HAV and 26 nonviral proteins with ≥2-fold isotope enrichment (o) in at least three of four independently labeled eHAV samples submitted for mass spectrometry. *Peptides of indeterminate human versus bovine origin (main text). †ALIX: PDCD6IP.
Fig. 3.
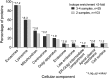
Cellular components mapped by FunRich (39) to proteins with ≥2-fold isotope enrichment in either ≥2 or ≥3 of 4 independently labeled eHAV samples. *−Log10(P value) for enrichment versus UniProt database.
Fig. S3.
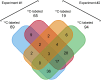
Venn diagram showing the distribution of viral and host cell proteins (n = 106) found to have ≥2-fold isotope enrichment in at least two of the four independently labeled eHAV samples submitted for mass spectrometry: one 13C-labeled (heavy) and one 12C-labeled (light) eHAV sample from each of two independent experiments. In experiment 1, eHAV particles produced in cells grown in the two different media were purified in iodixanol gradients before mixing, whereas in experiment 2 virus harvests were mixed before iodixanol gradient centrifugation (see SI Materials and Methods for details). The total number of proteins with ≥2-fold enrichment is shown below the label for each sample.
Fig. 4.
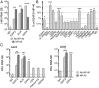
Immunoprecipitation of eHAV. (A) HAV RNA precipitated from gradient-purified eHAV preparations (mean ± SD) by anti-capsid (K24F2), anti-ALIX, or anti-DPP4 antibodies before (solid bars) and after (striped bars) Nonidet P-40 treatment. IgG, nonspecific antibody. (B) Antibody-mediated pull-downs of eHAV in the absence of detergent treatment. Results shown are mean percentage ± SD of the amount of HAV RNA precipitated by monoclonal K24F2 anti-HAV following Nonidet P-40 treatment. (C) eHAV precipitated by selected antibodies from two acute-phase infection human sera: (Left) A424 and (Right) A595. IgG, control Ig. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.5 for comparisons with control IgG by ANOVA with Dunn’s or Sidak’s tests for multiple comparisons; asterisks in parentheses indicate P values for comparisons between Nonidet P-40 treatment and no Nonidet P-40 by unpaired t test. n = 3 to 9 independent RT-PCR assays.
Fig. S4.
Lack of neutralization of eHAV infectivity by antibodies specific for eHAV surface-associated proteins by infrared immunofocus reduction assay. (A) Percent HAV antigen expression in cell cultures infected with antibody–eHAV mixtures as the percent of antigen expressed by cells infected with eHAV mixed with isotype control Ig. Data are mean ± range, n = 2 independent experiments. (B) Representative infrared fluorescence scans of HAV immunofoci developing in cell sheets infected with HM175/p16 eHAV–antibody mixtures. Shown (Top) is a control culture infected with virus mixed with the cognate isotype Ig control: rabbit for anti-DPP4, and murine for anti-CD9, anti-CD63, and anti-CD81.
Fig. 5.
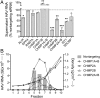
RNAi analysis of potential DPP4-, RAB7A-, and ESCRT-III–associated host protein function in eHAV biogenesis. (A) RT-PCR detection of virus released into supernatant fluids of persistently infected cell cultures between 72 and 96 h after transfection with host mRNA-specific or control, nontargeting siRNA pools. Results represent the percentage HAV RNA relative to that in supernatants from cultures transfected with nontargeting siRNA, and are shown as mean ± range from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Holm–Sidak’s test for multiple comparisons. (B) eHAV present in fractions of iodixanol gradients loaded with 96-h supernatants from cell cultures transfected with the indicated host mRNA-specific siRNAs. Nontargeting siRNA control results are shown as bars. Also shown are the density traces (dashed lines) from each gradient. eHAV was quantified by computing the area under the curve (AUC) for peaks in the RNA profiles.
Fig. S5.
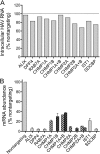
siRNA knockdown of host gene expression. (A) Impact of host mRNA-targeting siRNAs on intracellular HAV RNA abundance 96 h post transfection. Results shown are the mean ± range of duplicate RT-PCR assays from a representative experiment, and are presented as genome equivalents per cell-culture well. siRNAs were transfected into cells as single SMARTpools (GE Dharmacon) of four siRNAs, or in some cases as pairs of SMARTpools (CHMP1A and CHIMP1B, and CHIMP2A and CHIMP2B). (B) Knockdown efficiencies achieved with siRNAs targeting host mRNAs. Results shown represent the mean ± range of replicate assays from a representative experiment, and are presented as the percent of mRNA present in cells transfected with nontargeting siRNA.
Comment in
- Unconventional secretion of hepatitis A virus.
Kirkegaard K. Kirkegaard K. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):6653-6655. doi: 10.1073/pnas.1707142114. Epub 2017 Jun 12. Proc Natl Acad Sci U S A. 2017. PMID: 28607085 Free PMC article. No abstract available.
Similar articles
- Redundant Late Domain Functions of Tandem VP2 YPX3L Motifs in Nonlytic Cellular Egress of Quasi-enveloped Hepatitis A Virus.
González-López O, Rivera-Serrano EE, Hu F, Hensley L, McKnight KL, Ren J, Stuart DI, Fry EE, Lemon SM. González-López O, et al. J Virol. 2018 Nov 12;92(23):e01308-18. doi: 10.1128/JVI.01308-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232181 Free PMC article. - Nonlytic Quasi-Enveloped Hepatovirus Release Is Facilitated by pX Protein Interaction with the E3 Ubiquitin Ligase ITCH.
Shirasaki T, González-López O, McKnight KL, Xie L, Shiota T, Chen X, Feng H, Lemon SM. Shirasaki T, et al. J Virol. 2022 Nov 9;96(21):e0119522. doi: 10.1128/jvi.01195-22. Epub 2022 Oct 26. J Virol. 2022. PMID: 36286484 Free PMC article. - Nonlytic cellular release of hepatitis A virus requires dual capsid recruitment of the ESCRT-associated Bro1 domain proteins HD-PTP and ALIX.
Shirasaki T, Feng H, Duyvesteyn HME, Fusco WG, McKnight KL, Xie L, Boyce M, Kumar S, Barouch-Bentov R, González-López O, McNamara R, Wang L, Hertel-Wulff A, Chen X, Einav S, Duncan JA, Kapustina M, Fry EE, Stuart DI, Lemon SM. Shirasaki T, et al. PLoS Pathog. 2022 Aug 15;18(8):e1010543. doi: 10.1371/journal.ppat.1010543. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 35969644 Free PMC article. - Hepatitis A Virus Genome Organization and Replication Strategy.
McKnight KL, Lemon SM. McKnight KL, et al. Cold Spring Harb Perspect Med. 2018 Dec 3;8(12):a033480. doi: 10.1101/cshperspect.a033480. Cold Spring Harb Perspect Med. 2018. PMID: 29610147 Free PMC article. Review. - The Role of Exosome and the ESCRT Pathway on Enveloped Virus Infection.
Ju Y, Bai H, Ren L, Zhang L. Ju Y, et al. Int J Mol Sci. 2021 Aug 22;22(16):9060. doi: 10.3390/ijms22169060. Int J Mol Sci. 2021. PMID: 34445766 Free PMC article. Review.
Cited by
- Hijacking host extracellular vesicle machinery by hepatotropic viruses: current understandings and future prospects.
Chu YD, Chen MC, Yeh CT, Lai MW. Chu YD, et al. J Biomed Sci. 2024 Oct 5;31(1):97. doi: 10.1186/s12929-024-01063-0. J Biomed Sci. 2024. PMID: 39369194 Free PMC article. Review. - Coxsackievirus B3 Activates Macrophages Independently of CAR-Mediated Viral Entry.
Mohamud Y, Lin JC, Hwang SW, Bahreyni A, Wang ZC, Luo H. Mohamud Y, et al. Viruses. 2024 Sep 13;16(9):1456. doi: 10.3390/v16091456. Viruses. 2024. PMID: 39339932 Free PMC article. - Trick-or-Trap: Extracellular Vesicles and Viral Transmission.
Bou JV, Taguwa S, Matsuura Y. Bou JV, et al. Vaccines (Basel). 2023 Sep 27;11(10):1532. doi: 10.3390/vaccines11101532. Vaccines (Basel). 2023. PMID: 37896936 Free PMC article. Review. - Hepatoviruses promote very-long-chain fatty acid and sphingolipid synthesis for viral RNA replication and quasi-enveloped virus release.
Shiota T, Li Z, Chen GY, McKnight KL, Shirasaki T, Yonish B, Kim H, Fritch EJ, Sheahan TP, Muramatsu M, Kapustina M, Cameron CE, Li Y, Zhang Q, Lemon SM. Shiota T, et al. Sci Adv. 2023 Oct 20;9(42):eadj4198. doi: 10.1126/sciadv.adj4198. Epub 2023 Oct 20. Sci Adv. 2023. PMID: 37862421 Free PMC article. - Nonenveloped Avian Reoviruses Released with Small Extracellular Vesicles Are Highly Infectious.
Wang Z, He M, He H, Kilby K, Antueno R, Castle E, McMullen N, Qian Z, Zeev-Ben-Mordehai T, Duncan R, Pan C. Wang Z, et al. Viruses. 2023 Jul 23;15(7):1610. doi: 10.3390/v15071610. Viruses. 2023. PMID: 37515296 Free PMC article.
References
- Ehrenfeld E, Domingo E, Roos RP, editors. The Picornaviruses. ASM; Washington, DC: 2010.
- Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. Naked viruses that aren’t always naked: Quasi-enveloped agents of acute hepatitis. Annu Rev Virol. 2014;1:539–560. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous