Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP•NPLOC4•UFD1L is enhanced by a mutation that causes multisystem proteinopathy - PubMed (original) (raw)
Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP•NPLOC4•UFD1L is enhanced by a mutation that causes multisystem proteinopathy
Emily E Blythe et al. Proc Natl Acad Sci U S A. 2017.
Abstract
p97 is a "segregase" that plays a key role in numerous ubiquitin (Ub)-dependent pathways such as ER-associated degradation. It has been hypothesized that p97 extracts proteins from membranes or macromolecular complexes to enable their proteasomal degradation; however, the complex nature of p97 substrates has made it difficult to directly observe the fundamental basis for this activity. To address this issue, we developed a soluble p97 substrate-Ub-GFP modified with K48-linked ubiquitin chains-for in vitro p97 activity assays. We demonstrate that WT p97 can unfold proteins and that this activity is dependent on the p97 adaptor NPLOC4-UFD1L, ATP hydrolysis, and substrate ubiquitination, with branched chains providing maximal stimulation. Furthermore, we show that a p97 mutant that causes inclusion body myopathy, Paget's disease of bone, and frontotemporal dementia in humans unfolds substrate faster, suggesting that excess activity may underlie pathogenesis. This work overcomes a significant barrier in the study of p97 and will allow the future dissection of p97 mechanism at a level of detail previously unattainable.
Keywords: AAA ATPase; Cdc48; Npl4; Proteasome; Ufd1.
Conflict of interest statement
Conflict of interest statement: R.J.D. is a founder and a shareholder of Cleave Biosciences, which is developing CB-5083 for therapy of cancer. The other authors declare that no competing interests exist.
Figures
Fig. S1.
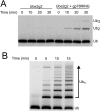
Characterization of gp78RING-Ube2g2 chimera. Time courses comparing the rates of ubiquitin polymerization by 0.1 µM Ube2g2 alone or in the presence of 20 µM gp78RING (A) or when Ube2g2 is fused directly to gp78RING (B; assayed at 0.1 µM). The chimera is more than sixfold faster than the unfused proteins.
Fig. 1.
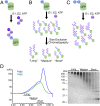
Substrate design and synthesis. (A) Preassembled, K48-linked Ub3 chains containing a K48R mutation on the distal ubiquitin were ligated onto a noncleavable linear His6-Ub-GFP fusion protein to produce pure Ub3Ub-GFP. (B) E1, E2, ubiquitin, and ATP were added to His6-Ub-GFP to elongate K48-linked ubiquitin chains of varying lengths on the ubiquitin fused to GFP. These resulting substrates were purified from free ubiquitin chains via Ni-NTA resin and crudely fractionated according to chain length via size-exclusion chromatography to produce pools of “long-”, “medium-”, and “short”-chain substrates (UbLUb-GFP, UbMUb-GFP, and UbSUb-GFP, respectively). (C) To produce branched chains, ubiquitin chains of varying length were enzymatically elongated on a di- or triubiquitin linear fusion protein, Ub-Ub-GFP, or Ub-Ub-Ub-GFP, similar to B. (D) Size-exclusion chromatogram and corresponding SDS/PAGE gel for the purification of substrate described in B.
Fig. 2.
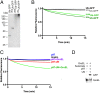
p97 unfolds Ub-GFP in a UN-dependent manner (A) SDS/PAGE analysis of GFP substrates with different ubiquitin chain structures stained with Coomassie Brilliant Blue. (B) Upon addition of ATP, 75 nM p97, 150 nM UN, and 250 nM GroEL, 25 nM Ub-GFP, and Ub3Ub-GFP did not appreciably lose fluorescence over time. However, Ub-GFP with “medium” or “long” K48-linked chains (UbMUb-GFP and UbLUb-GFP) exhibited 26% and 32% loss of signal after 15 min, respectively. Representative traces shown (n ≥ 3). (C) Fluorescence of UbLUb-GFP did not change over time with the addition of p97, GroEL, or p97 plus GroEL. Upon addition of p97 plus UN, a small decrease in signal was observed, and this decrease was augmented with the addition of GroEL. Representative traces shown (n ≥ 2). (D) UbLUb-GFP coimmunoprecipitated with GroEL only in the presence of p97 and UN.
Fig. S2.
SDS/PAGE analysis of proteins used in this study.
Fig. S3.
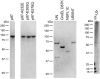
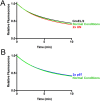
p97 adaptors NSFL1C and UBXN7 do not promote substrate processing. (A) In the presence of p97 and GroEL, UN promoted a decrease in fluorescence of UbLUb-UbLUb-GFP, indicating substrate unfolding. However, NSFL1C and UBXN7 were not able to replace UN. Representative traces shown (n ≥ 2). (B) NSFL1C and UBXN7 bind UbLUb-UbLUb-GFP and mediate its interaction with p97, as shown by co-IP with purified proteins. Reactions contained 100 nM p97, 200 nM adaptor, and 200 nM substrate and were immunoprecipitated with antibodies to GFP or p97 as indicated.
Fig. S4.
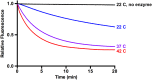
Unfolding by p97 is temperature-dependent. Unfolding reactions using UbLUb-UbLUb-GFP were carried out at 22 °C, 37 °C, and 42 °C. Higher temperatures increased the rate and extent of the unfolding reaction. Representative traces shown (n ≥ 2).
Fig. 3.
Branched ubiquitin chains are better p97 substrates. (A) SDS/PAGE analysis of Ub-GFP substrates stained with Coomassie Brilliant Blue. (B) Comparison of unfolding of substrates with one, two, or three ubiquitins fused in tandem to GFP. Note that substrate with two linearly fused ubiquitins (e.g., UbMUb-UbMUb-GFP) was unfolded to a greater extent by p97 than substrate with a single linearly fused ubiquitin even though aggregate ubiquitination for the latter substrate was at least as extensive or greater than the former. Addition of an additional linearly fused ubiquitin (UbLUb-UbLUb-UbLUb–GFP) yielded no further improvement. Representative traces shown (n ≥ 3).
Fig. S5.
Unfolding reaction components are saturating. (A) When the concentration of GroEL was halved or the concentration of UN was doubled, minimal changes in the rate of unfolding of UbLUb-UbLUb-GFP were observed. Representative traces shown (n = 2). (B) When the concentration of p97 was doubled, virtually no change in the rate of unfolding of UbLUb-UbLUb-GFP was observed. Representative traces shown (n = 2).
Fig. S6.
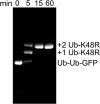
Two Ub-K48R modifications can be added onto Ub-Ub-GFP. Time course of reaction of 10 µM Ub-Ub-GFP with 200 µM Ub-K48R under conditions identical to those used to make substrate produced pure Ub1Ub-Ub1Ub-GFP, indicating that ubiquitin chains are assembled on both fused ubiquitins in Ub-Ub-GFP. SDS/PAGE analysis of unboiled samples. GFP was detected by excitation at 488 nm.
Fig. 4.
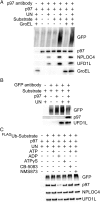
ATPase activity of p97 is critical for and stimulated by substrate unfolding. (A) Fluorescence traces of UbLUb-UbLUb-GFP in the presence of p97, UN, GroEL, and various nucleotides and p97 inhibitors. Unfolding was observed only in the presence of ATP. Representative traces shown (n ≥ 2). (B) Unfolding of UbLUb-UbLUb-GFP by p97 ATPase mutants. p97-E305Q and p97-E578Q are deficient in D1 and D2 ATPase activity, respectively. p97-E305Q was able to unfold substrate, whereas p97-E578Q was not. Representative traces shown (n ≥ 3). (C) Substrate stimulates ATPase activity of p97 when UN is present. Unanchored ubiquitin chains and those linked to Ub-GFP yielded equivalent stimulation, whereas Ub-GFP and Ub3Ub-GFP did not stimulate. ATPase activity was measured with BIOMOL Green as described in Methods and was normalized to basal WT p97 activity. Error bars represent SD (n = 4). (D) Effect of substrate plus UN on ATPase activity of D1 and D2 domain ATPase mutants. Addition of UbLUb-UbLUb-GFP plus UN slightly decreased ATPase activities of D1 mutant E305Q and D2 mutant E578Q. Error bars represent SD (n = 4).
Fig. S7.
Substrate alone or in complex with other adaptors does not accelerate p97 ATPase. (A) ATPase activity of p97 is not affected by substrate when UN is absent. (B) NSFL1C and UBXN7 do not promote substrate-induced ATPase acceleration. Addition of substrate did not significantly change the ATPase activity of p97•NSFL1C or p97•UBXN7. ATPase activity was measured with BIOMOL Green as described in Methods. Error bars represent SD (n = 4).
Fig. 5.
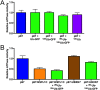
UN recruits ubiquitylated substrate to p97. (A) Association of substrate with p97 depends on UN. Reactions containing 100 nM p97, 200 nM UN, 200 nMUbLUb-UbLUb-GFP, and/or 200 nM GroEL were immunoprecipitated by using an anti-p97 antibody and assessed by Western blot. Substrate was pulled down with p97 only in the presence of UN, and substrate enhanced the binding of UN to p97. Effects of substrate and UN on binding of GroEL to p97 could not be determined as a result of high background binding of GroEL to beads, antibody, and/or p97. (B) UN binds directly to substrate and links it to p97. Samples prepared as in A were immunoprecipitated with an anti-GFP antibody. Substrate pulled down UN but bound p97 only in the presence of UN. (C) Bound nucleotide has only a modest effect on formation of a p97•UN•substrate ternary complex. Samples prepared as in A with various nucleotides and/or p97 inhibitors were immunoprecipitated by anti-FLAG resin, which bound the FLAG-tagged ubiquitin on substrate. Binding of p97, but not UN, was reduced only in the ADP state.
Fig. 6.
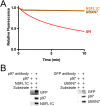
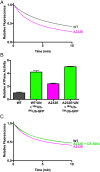
IMBPFD mutant p97-A232E is a better unfoldase. (A) In the presence of UN and GroEL, p97-A232E catalyzed the loss of fluorescence of UbLUb-UbLUb-GFP faster than WT, suggesting it acts as an improved unfoldase. Rates are listed in Table 1, and the difference between WT and p97-A232E is statistically significant at P < 0.0001. Representative traces shown (n ≥ 3). (B) p97-A232E shows accelerated ATPase rates in the presence of UN and substrate. The IBMPFD mutant also had a higher baseline ATPase rate than WT (unpaired t test, P < 0.0001) and a higher ATPase activity in the presence of UN and substrate compared with WT (unpaired t test, P = 0.0003). ATPase activity was measured with BIOMOL Green as described in Methods and was normalized to basal WT p97 activity. Error bars represent SD (n = 4). (C) Addition of a 37.5-nM CB-5083 restores p97-A232E unfoldase activity to WT levels. Representative traces shown (n = 2), and the difference between WT and p97-A232E rates is statistically significant (unpaired t test, P = 0.02). Independent preparations of proteins were used for A and C.
Similar articles
- A unique IBMPFD-related P97/VCP mutation with differential binding pattern and subcellular localization.
Erzurumlu Y, Kose FA, Gozen O, Gozuacik D, Toth EA, Ballar P. Erzurumlu Y, et al. Int J Biochem Cell Biol. 2013 Apr;45(4):773-82. doi: 10.1016/j.biocel.2013.01.006. Epub 2013 Jan 16. Int J Biochem Cell Biol. 2013. PMID: 23333620 - Valosin-Containing Protein (VCP)/p97 Oligomerization.
Yu G, Bai Y, Zhang ZY. Yu G, et al. Subcell Biochem. 2024;104:485-501. doi: 10.1007/978-3-031-58843-3_18. Subcell Biochem. 2024. PMID: 38963497 Review. - Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4·UFD1L Binding and Substrate Processing.
Blythe EE, Gates SN, Deshaies RJ, Martin A. Blythe EE, et al. Structure. 2019 Dec 3;27(12):1820-1829.e4. doi: 10.1016/j.str.2019.09.011. Epub 2019 Oct 14. Structure. 2019. PMID: 31623962 Free PMC article. - Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Nandi P, Li S, Columbres RCA, Wang F, Williams DR, Poh YP, Chou TF, Chiu PL. Nandi P, et al. Int J Mol Sci. 2021 Jul 28;22(15):8079. doi: 10.3390/ijms22158079. Int J Mol Sci. 2021. PMID: 34360842 Free PMC article. - The Cure VCP Scientific Conference 2021: Molecular and clinical insights into neurodegeneration and myopathy linked to multisystem proteinopathy-1 (MSP-1).
Johnson MA, Klickstein JA, Khanna R, Gou Y; Cure VCP Disease Research Consortium; Raman M. Johnson MA, et al. Neurobiol Dis. 2022 Jul;169:105722. doi: 10.1016/j.nbd.2022.105722. Epub 2022 Apr 8. Neurobiol Dis. 2022. PMID: 35405261 Free PMC article. Review.
Cited by
- The Cryo-EM Effect: Structural Biology of Neurodegenerative Disease Proteostasis Factors.
Creekmore BC, Chang YW, Lee EB. Creekmore BC, et al. J Neuropathol Exp Neurol. 2021 Jun 4;80(6):494-513. doi: 10.1093/jnen/nlab029. J Neuropathol Exp Neurol. 2021. PMID: 33860329 Free PMC article. Review. - Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau.
Darwich NF, Phan JM, Kim B, Suh E, Papatriantafyllou JD, Changolkar L, Nguyen AT, O'Rourke CM, He Z, Porta S, Gibbons GS, Luk KC, Papageorgiou SG, Grossman M, Massimo L, Irwin DJ, McMillan CT, Nasrallah IM, Toro C, Aguirre GK, Van Deerlin VM, Lee EB. Darwich NF, et al. Science. 2020 Nov 20;370(6519):eaay8826. doi: 10.1126/science.aay8826. Epub 2020 Oct 1. Science. 2020. PMID: 33004675 Free PMC article. - Localized K63 ubiquitin signaling is regulated by VCP/p97 during oxidative stress.
Maduka AO, Manohar S, Foster MW, Silva GM. Maduka AO, et al. bioRxiv [Preprint]. 2024 Jun 21:2024.06.20.598218. doi: 10.1101/2024.06.20.598218. bioRxiv. 2024. PMID: 38948861 Free PMC article. Preprint. - A Futile Battle? Protein Quality Control and the Stress of Aging.
Higuchi-Sanabria R, Frankino PA, Paul JW 3rd, Tronnes SU, Dillin A. Higuchi-Sanabria R, et al. Dev Cell. 2018 Jan 22;44(2):139-163. doi: 10.1016/j.devcel.2017.12.020. Dev Cell. 2018. PMID: 29401418 Free PMC article. Review. - Valosin-Containing Protein (VCP): A Review of Its Diverse Molecular Functions and Clinical Phenotypes.
Pontifex CS, Zaman M, Fanganiello RD, Shutt TE, Pfeffer G. Pontifex CS, et al. Int J Mol Sci. 2024 May 22;25(11):5633. doi: 10.3390/ijms25115633. Int J Mol Sci. 2024. PMID: 38891822 Free PMC article. Review.
References
- Wang Q, Song C, Li C-CH. Molecular perspectives on p97-VCP: Progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146:44–57. - PubMed
- Wolf DH, Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta. 2012;1823:117–124. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous