w09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS-RAF1-MAP2K-MAPK1/3 pathway - PubMed (original) (raw)
. 2017 Jul 3;13(7):1093-1112.
doi: 10.1080/15548627.2017.1319039. Epub 2017 May 17.
Zuguo Zheng 1, Li Ling 1, Xiaohui Yang 2, Ni Zhang 1, Xue Wang 1, Maozhi Hu 3, Yu Xia 1, Yiwen Ma 1, Haoran Yang 1, Yunyi Wang 1, Hongqi Liu 4
Affiliations
- PMID: 28513279
- PMCID: PMC5529067
- DOI: 10.1080/15548627.2017.1319039
w09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS-RAF1-MAP2K-MAPK1/3 pathway
Pinghu Zhang et al. Autophagy. 2017.
Abstract
The EGFR (epidermal growth factor receptor) signaling pathway is frequently deregulated in many malignancies. Therefore, targeting the EGFR pathway is regarded as a promising strategy for anticancer drug discovery. Herein, we identified a 2-amino-nicotinonitrile compound (w09) as a novel autophagy enhancer, which potently induced macroautophagy/autophagy and consequent apoptosis in gastric cancer cells. Mechanistic studies revealed that EGFR-mediated activation of the RAS-RAF1-MAP2K-MAPK1/3 signaling pathway played a critical role in w09-induced autophagy and apoptosis of gastric cancer cells. Inhibition of the MAPK1/3 pathway with U0126 or blockade of autophagy by specific chemical inhibitors markedly attenuated the effect of w09-mediated growth inhibition and caspase-dependent apoptosis. Furthermore, these conclusions were supported by knockdown of ATG5 or knockout of ATG5 and/or ATG7. Notably, w09 increased the expression of SQSTM1 by transcription, and knockout of SQSTM1 or deleting the LC3-interaction region domain of SQSTM1, significantly inhibited w09-induced PARP1 cleavage, suggesting the central role played by SQSTM1 in w09-induced apoptosis. In addition, in vivo administration of w09 effectively inhibited tumor growth of SGC-7901 xenografts. Hence, our findings not only suggested that activation of the EGFR-RAS-RAF1-MAP2K-MAPK1/3 signaling pathway may play a critical role in w09-induced autophagy and apoptosis, but also imply that induction of autophagic cancer cell death through activation of the EGFR pathway may be a potential therapeutic strategy for EGFR-disregulated gastric tumors.
Keywords: ATG7; EGFR; LC3; RAS-RAF1-MAP2K-MAPK1/3; SQSTM1; apoptosis; autophagy; gastric cancer; lysosome; w09.
Figures
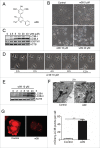
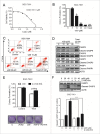
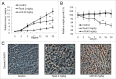
Figure 1.
w09 induces autophagy in gastric cancer cells. (A) The structure of compound w09. (B) SGC-7901 cells were treated with the indicated concentrations of w09 or vehicle (DMSO) for 12 h, representative images of SGC-7901 cells are shown as photographed with a phase contrast microscope (× 200); scale bar: 20 µm. (C) The expression of SQSTM1 and conversion of LC3B-I to LC3B-II were evaluated by immunoblotting. (D) SGC-7901 cells were exposed to w09 (10 μM) for the indicated time, and representative images of SGC-7901 cells are shown (× 400); scale bar: 10 μm. (E) Conversion of LC3B-I to LC3B-II was analyzed by immunoblotting. (F) SGC-7901 cells were treated with 10 μM w09 for 6 h. Representative microscopy images of SGC-7901 cells were obtained by transmission electron microscopy. The black arrow indicates autophagic vacuoles containing cytoplasmic context. Scale bars: 0.5 μm. (G) Representative confocal fluorescence images of SGC-7901 cells stably expressing mCherry-LC3B, treated with w09 or vehicle for 6 h. The difference of mCherry-LC3 puncta in cells treated with w09 or vehicle was quantified with ImageJ and statistical analysis was conducted using the Student t test. **P<0.01.
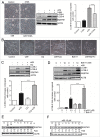
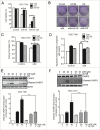
Figure 2.
Blockage of autophagy prevents w09-induced autophagy in gastric cancer cells. (A) SGC-7901 cells were pretreated with 3-MA (10 mM) for 1 h, followed by treatment with w09 (10 μM) or vehicle (DMSO) for an additional 6 h. Representative microscopy images of SGC-7901 cells were obtained by phase contrast microscopy (× 200); scale bar 20 μm. The expression of autophagy-related proteins, LCB3-I, LC3B-II and SQSTM1 was assessed by immunoblotting. Quantification represents the relative protein levels of the cells normalized to ACTB (mean ± SD from 3 independent experiments (***P<0.001). (B) SGC-7901 cells were pretreated with CQ (20 μM) or BafA1 (1 or 10 nM) for 1 h, followed by treatment with w09 (10 μM) and vehicle (DMSO) for an additional 6 h and representative microscopy images of SGC-7901 cells were obtained by phase contrast microscopy (× 200); scale bar: 20 μm. (C) and (D) The expression of autophagy-related proteins, LC3B-I, LC3B-II and SQSTM1 of SGC-7901 cells treated as described in (B) was assessed by immunoblotting. Quantification represents the relative protein levels of the cells normalized to ACTB (Mean ± SD from 3 independent experiments (***P<0.001). (E) and (F) SGC-7901 cells were treated with w09 (10 μM) or CQ (20 μM) for 0 to 12 h and the expression of SQSTM1 protein was analyzed by western blot. SQSTM1 fold-increase levels compared with “0 h” are shown.
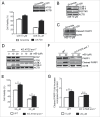
Figure 3.
w09 induces autophagic flux in gastric cancer cells. HeLa (A) or SGC-7901 (B) cells stably expressing mCherry-LC3 were treated with the indicated drugs for 6 h, and representative fluorescent images (× 200) were photographed by laser scaning confocal microscopy (Olympus FV1000, Tokyo, Japan); scale bar: 20 μm. The difference of mCherry-LC3 dots in HeLa (C) and SGC-7901 (D) cells was quantified with ImageJ and at least 20 cells of each treatment were used to calculate statistical significance using the Student t test (**p<0.01) as described previously.
Figure 4.
w09 induces autophagy via activation of the EGFR-RAS-RAF1-MAP2K-MAPK1/3 signaling pathway. (A) SGC-7901 cells were pretreated with 10 μM U0126 for 1 h, followed by coincubation with w09 (10 μM) or vehicle (DMSO) for 12 h. Representative microscopy images of SGC-7901 cells are shown (× 200); scale bar: 50 μm. (B) The expression of the autophagy-associated protein and autopahagosome marker LC3B-II was analyzed by western blot, and its relative levels, normalized to ACTB (mean ± S.D) from 3 independent experiments (**P<0.01) were quantified (C). (D) The phosphorylation levels of the MAP2K-MAPK1/3 signaling pathway in cells treated with w09 were detected by immunoblot analysis. (E) SGC-7901 cells were pretreated with sorafenib (20 μM) for 1 h, followed by coincubation with w09 (10 μM) or vehicle (DMSO). The phosphorylation levels of the MAP2K-MAPK1/3 signaling pathway were assessed by immunoblot analysis, and (F) the expression levels of LC3B-I/LC3B-II were also detected by immunoblot analysis. The quantification of the LC3B-II/ACTB ratio is presented (***P<0.001). (G) SGC-7901 cells were pretreated with gefitinib (20 μM) for 1 h, followed by coincubation with w09 (10 μM) or vehicle (DMSO), the phosphorylation levels of the MAP2K-MAPK1/3 signaling pathway of SGC-7901 cells were assessed by immunoblot analysis, and (H) the expression levels of LC3B-I:LC3B-II were also detected by immunoblot analysis. The quantification of LC3B-II/ACTB ratio is presented (**P<0.01). After SGC-7901 cells were treated with various concentrations of w09 for 2 h (I), or inoculated with 10 μM w09 for different time intervals as indicated (J), the phosphorylation of the EGFR-RAS-RAF1-MAP2K-MAPK1/3 signaling pathway was detected by western blot.
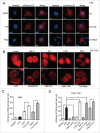
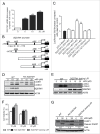
Figure 5.
EGFR plays a critical role in the regulation of w09-induced autophagy. (A) SGC-7901 cells were exposed to w09 or EGF for 6 h and the activation effect of w09 or EGF on the MAPK1/3 signaling pathway was assessed by western blot. (B) SGC-7901 cells were pretreated with EGF for 2 h, followed by incubation with w09 for 6 h and representative microscopy images of SGC-7901 cells treated as described is shown (× 200); scale bar: 20 μm. (C) SGC-7901 cells were treated with w09 alone or combined with EGF for 6 h. The conversion of LC3B-I to LC3B-II was assessed by western blot and relative fold-increase of LC3B-II is presented. (D) Expression of EGFR in the _egfr_-null CHO-K1 and CHO-K1 cells transfected with an EGFR-expression plasmid was examined by immunoblot analysis. (E) _egfr_-null CHO-K1 cells and _Egfr_-transfected CHO-K1 cells were treated with the indicated concentrations of w09 for 6 h. Representative microscopy images of SGC-7901 cells were obtained (× 200; scale bar: 20 μm) and the expression of LC3B-II was determined by immunoblot analysis (F).
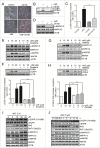
Figure 6.
w09 inhibits gastric cancer cell growth through the CASP8-dependent apoptosis pathway. (A) SGC-7901 cells were treated with various concentrations of w09 for 72 h. Cell viability was assessed by MTT assay. (B) SGC-7901 cells were treated with various concentrations of w09 for 3 d, followed by incubation with new medium without w09 for an additional 9 d. Cell colonies were stained with crystal violet and quantification of the colonies (cell number >20) normalized to control is presented (mean ± SD for 3 independent experiments, *P<0.05, and ***_P_<0.001). (C) SGC-7901 cells were treated with w09 or vehicle (DMSO) for 48 h and apoptosis was analyzed by FACS using an ANXA5-EGFP and PI cell apoptosis kit. Representative results from 3 independent experiments are presented. (D) Cells were treated as in (C), total protein was prepared and the expression of the apoptosis-related proteins was analyzed by western blot using antibodies against total or cleaved CASP9, CASP8, CASP3 and PARP1. (E) SGC-7901 cells were pretreated with a pan-caspase inhibitor, Z-VAD-fmk (10 μM) for 1 h, followed by incubation with 20 μM of w09 or vehicle (DMSO) for 2 more d and then cells were cultured in absence of w09 for an additional 10 d. Cell colonies were stained with crystal violet, and quantification of the colonies (cell number >50) normalized to control is presented (mean ± SD for 3 independent experiments; **P<0.01). (F) SGC-7901 cells were preincubated with Z-VAD-fmk (10 μM) for 1 h, followed by treatment with the indicated concentrations of w09 for an additional 48 h. Whole-cell lysates were prepared for western blot analysis of cleaved-CASP3 and quantification of the cleaved CASP3 levels is shown (mean ± SD from 3 independent experiments; **P<0.01 and ***P<0.001).
Figure 7.
Blocking w09-induced autophagy significantly attenuates w09-mediated apoptosis in gastric cancer cells. (A) SGC-7901 cells were pretreated with CQ (20 μM) or U0126 (10 μM) for 1 h, followed by coincubation with various concentrations of w09 for an additional 72 h. Cell viability was assessed by MTT assay (**P<0.01 vs control). (B) SGC-7901 cells were pretreated with CQ (20 μM) or U0126 (10 μM) for 1 h, followed by coincubation with 20 μM w09 for 48 h and then cells were cultured in the absence of w09 for an additional 10 d. Cell colonies were stained with crystal violet and (C) quantification of the colonies (cell number >50) normalized to control in (B) is presented (mean ± SD for 3 independent experiments; **P<0.01). (D) SGC-7901 cells were pretreated with CQ (20 μM) or U0126 (10 μM) for 1 h, followed by coincubation with 20 μM w09 for 48 h. Cell apoptosis analysis was performed by FACS using ANXA5-EGFP and PI cell apoptosis kit. Quantification of apoptosis in cells treated as described in (D) is presented (mean ± SD for duplicate experiments; **P<0.01). ((E)and F) Total proteins of SGC-7901 cells treated as described in (D) were extracted and the expression of cleaved CASP3 was detected by western blot analysis, and quantification of cleaved CASP3 is presented as mean ± SD from 3 independent experiments (*** P<0.001).
Figure 8.
Knockdown or knockout of autophagy-related genes markedly prevents w09-induced autophagy and apoptosis in gastric cancer cells. (A) SGC-7901 cells were transfected with ATG5 or control (Scrambled) siRNA for 48 h and the expression of ATG5 was evaluated by western blot (upper panel). 48 h after transfection, SGC-7901 cells were treated with w09 for 48 h and cell viability was assessed by MTT assay (lower panel) (**P<0.01). (B) SGC-7901 cells were transfected with ATG5 or control (Scrambled) siRNA for 48 h and then treated with 10 μM w09 for 6 h, the conversion of LC3B-I to LC3B-II and the expression of SQSTM1 was evaluated by western blot. (C) SGC-7901 cells were transfected with ATG5 or control (Scrambled) siRNA for 48 h and then treated with 20 μM w09 for 24 h. Cleaved CASP3 in cells was analyzed by western blot. (D) A wild-type SGC-7901 cell line and a new SGC-7901 cell line with double knockout of ATG5 and ATG7 genes were treated with w09 for 6 h, the expression of ATG5 and ATG7, as well as the conversion of LC3B-I to LC3B-II were assessed by western blot. (E) Wild-type SGC-7901 cells or SGC-7901 cells with ATG5 ATG7 double-knockout were treated with w09 for 48 h. Cell viability was evaluated by MTT assay (**P<0.01) and (F) the cleavage of PARP1 was analyzed with western blot and quantified (G).
Figure 9.
SQSTM1 plays an important role in w09-mediated cell apoptosis in gastric cancer cells. (A), w09 promotes SQSTM1 mRNA transcript in a dose-dependent manner. SGC-7901 cells were treated with w09 for 12 h with the indicated dose of w09 and the relative level of SQSTM1 mRNA of cells in the absence or presence of w09 was analyzed by real-time PCR. ((B)and C), Reporter gene assays were performed using wild-type (−1781/+46) or the indicated deleted or mutated SQSTM1 promoter constructs as described previously., SGC-7901 cells were transfected with an empty vector (pGL3-Basic) or the indicated SQSTM1 promoter constructs. Cells were harvested 24 h after transfection and the relative promoter activities are expressed as the ratio between measured luciferase and renilla plasmid activities. The data shown are the mean activities obtained in one experiment preformed in triplicate (**P<0.01). A knockout of SQSTM1 in the SGC-7901 cell line (D) and a SGC-7901 cell line lacking the SQSTM1 LIR domain (E) were constructed and the autophagic effect of w09 on these 2 new cell lines was confirmed by western blot. All these cell lines were treated with w09 for 6 h and the autophagic markers LC3 and SQSTM1 were analyzed with western blot. (F) Compared with wild type, knockout of SQSTM1 or lack of the SQSTM1 LIR domain significantly rescued cell death-induced by w09 and (G) significantly inhibited the cleavage of PARP1 induced by w09. All these cell lines were treated with w09 for 48 h. The inhibitory effect of w09 was analyzed with MTT assay and the apoptotic effect of w09 was examined with western blot. Relative fold-increase of cleaved PARP1 compared with the negative control (DMSO) is presented.
Figure 10.
w09 inhibits tumor growth in an SGC-7901 xenograft model. (A) Relative tumor volume in vehicle control mice, w09-treated mice and taxel-treated mice. Error bars represent means ± SD **P<0.01 indicates significant difference in relative tumor volume compared with control, w09 or taxel. (B) Relative weight changes of mice during 21 d of exposure. Error bars represent means ± SD **P<0.01 indicates significant difference in relative body weight growth of the indicated treatment group. (C) Paraffin-embedded tumor tissue sections were stained with hematoxylin and eosin staining and an antibody against cleaved CASP3 for evaluating apoptosis was used. Representative photos are presented (× 200); scale bar: 20 μm.
Similar articles
- ATG7-dependent and independent autophagy determine the type of treatment in lung cancer.
Zhang P, Ling L, Zheng Z, Zhang Y, Wang R, Wu M, Zhang N, Hu M, Yang X. Zhang P, et al. Pharmacol Res. 2021 Jan;163:105324. doi: 10.1016/j.phrs.2020.105324. Epub 2020 Dec 1. Pharmacol Res. 2021. PMID: 33276100 - A specific super-enhancer actuated by berberine regulates EGFR-mediated RAS-RAF1-MEK1/2-ERK1/2 pathway to induce nasopharyngeal carcinoma autophagy.
Wu Y, Jia Q, Tang Q, Chen L, Deng H, He Y, Tang F. Wu Y, et al. Cell Mol Biol Lett. 2024 Jun 28;29(1):92. doi: 10.1186/s11658-024-00607-4. Cell Mol Biol Lett. 2024. PMID: 38943090 Free PMC article. - BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells.
Goulielmaki M, Koustas E, Moysidou E, Vlassi M, Sasazuki T, Shirasawa S, Zografos G, Oikonomou E, Pintzas A. Goulielmaki M, et al. Oncotarget. 2016 Feb 23;7(8):9188-221. doi: 10.18632/oncotarget.6942. Oncotarget. 2016. PMID: 26802026 Free PMC article. - Targeting EGFR-mediated autophagy as a potential strategy for cancer therapy.
Sooro MA, Zhang N, Zhang P. Sooro MA, et al. Int J Cancer. 2018 Nov 1;143(9):2116-2125. doi: 10.1002/ijc.31398. Epub 2018 Apr 6. Int J Cancer. 2018. PMID: 29574749 Review. - Revisiting autophagy addiction of tumor cells.
Nyfeler B, Eng CH. Nyfeler B, et al. Autophagy. 2016 Jul 2;12(7):1206-7. doi: 10.1080/15548627.2016.1170265. Epub 2016 Apr 20. Autophagy. 2016. PMID: 27097231 Free PMC article. Review.
Cited by
- Nintedanib Induces the Autophagy-Dependent Death of Gastric Cancer Cells by Inhibiting the STAT3/Beclin1 Pathway.
Zhu H, Xia MM, Tong KH, Duan WB. Zhu H, et al. Dig Dis Sci. 2023 Apr;68(4):1280-1291. doi: 10.1007/s10620-022-07653-y. Epub 2022 Aug 24. Dig Dis Sci. 2023. PMID: 36002676 - Autophagic Regulation of p62 is Critical for Cancer Therapy.
Islam MA, Sooro MA, Zhang P. Islam MA, et al. Int J Mol Sci. 2018 May 8;19(5):1405. doi: 10.3390/ijms19051405. Int J Mol Sci. 2018. PMID: 29738493 Free PMC article. Review. - Comprehensive Analysis of E2F Family Members in Human Gastric Cancer.
Li S, Yang X, Li W, Chen Z. Li S, et al. Front Oncol. 2021 Aug 31;11:625257. doi: 10.3389/fonc.2021.625257. eCollection 2021. Front Oncol. 2021. PMID: 34532281 Free PMC article. - TRIM65 Promotes Cervical Cancer Through Selectively Degrading p53-Mediated Inhibition of Autophagy and Apoptosis.
Wang XY, Mao HW, Guan XH, Huang QM, Yu ZP, Wu J, Tan HL, Zhang F, Huang X, Deng KY, Xin HB. Wang XY, et al. Front Oncol. 2022 Mar 24;12:853935. doi: 10.3389/fonc.2022.853935. eCollection 2022. Front Oncol. 2022. PMID: 35402260 Free PMC article. - Quercetin Attenuates Podocyte Apoptosis of Diabetic Nephropathy Through Targeting EGFR Signaling.
Liu Y, Li Y, Xu L, Shi J, Yu X, Wang X, Li X, Jiang H, Yang T, Yin X, Du L, Lu Q. Liu Y, et al. Front Pharmacol. 2022 Jan 5;12:792777. doi: 10.3389/fphar.2021.792777. eCollection 2021. Front Pharmacol. 2022. PMID: 35069207 Free PMC article.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855 - PubMed
- Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget 2015; 6:24750-24779; PMID:26267324; https://doi.org/10.18632/oncotarget.4990 - DOI - PMC - PubMed
- Xu YH, Richert N, Ito S, Merlino GT, Pastan I. Characterization of epidermal growth factor receptor gene expression in malignant and normal human cell lines. Proc Natl Acad Sci USA 1984; 81:7308-7312; PMID:6095284; https://doi.org/10.1073/pnas.81.23.7308 - DOI - PMC - PubMed
- Hirsch FR, Varella-Garcia M, Bunn PA Jr, Di Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol 2003; 21:3798-3807; PMID:12953099; https://doi.org/10.1200/JCO.2003.11.069 - DOI - PubMed
- Shia J, Klimstra DS, Li AR, Qin J, Saltz L, Teruya-Feldstein J, Akram M, Chung KY, Yao D, Paty PB, et al.. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol 2005; 18:1350-1356; PMID:15832190; https://doi.org/10.1038/modpathol.3800417 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous