Computational Exploration of Putative LuxR Solos in Archaea and Their Functional Implications in Quorum Sensing - PubMed (original) (raw)
Computational Exploration of Putative LuxR Solos in Archaea and Their Functional Implications in Quorum Sensing
Akanksha Rajput et al. Front Microbiol. 2017.
Abstract
LuxR solos are unexplored in Archaea, despite their vital role in the bacterial regulatory network. They assist bacteria in perceiving acyl homoserine lactones (AHLs) and/or non-AHLs signaling molecules for establishing intraspecies, interspecies, and interkingdom communication. In this study, we explored the potential LuxR solos of Archaea from InterPro v62.0 meta-database employing taxonomic, probable function, distribution, and evolutionary aspects to decipher their role in quorum sensing (QS). Our bioinformatics analyses showed that putative LuxR solos of Archaea shared few conserved domains with bacterial LuxR despite having less similarity within proteins. Functional characterization revealed their ability to bind various AHLs and/or non-AHLs signaling molecules that involve in QS cascades alike bacteria. Further, the phylogenetic study indicates that Archaeal LuxR solos (with less substitution per site) evolved divergently from bacteria and share distant homology along with instances of horizontal gene transfer. Moreover, Archaea possessing putative LuxR solos, exhibit the correlation between taxonomy and ecological niche despite being the inhabitant of diverse habitats like halophilic, thermophilic, barophilic, methanogenic, and chemolithotrophic. Therefore, this study would shed light in deciphering the role of the putative LuxR solos of Archaea to adapt varied habitats via multilevel communication with other organisms using QS.
Keywords: Archaea; LuxR solos; bioinformatics analyses; ecological niche; extremophiles; ligand-binding; phylogeny; quorum-sensing.
Figures
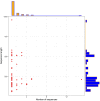
FIGURE 1
Archaea LuxR containing sequences statistics. Scattered plot with the marginal histogram showing a correlation between sequence length and the number of sequences.
FIGURE 2
Distribution of unique domains in 110 LuxR containing archaea sequences extracted InterPro. [_IPR000792, Transcriptional regulator LuxR, C-terminal domain; IPR007050, Bacterioopsin activator-type, HTH domain; IPR013324, RNA polymerase sigma factor, region 3/4; IPR003018, GAF domain; IPR031803, Bacterioopsin transcriptional activator, GAF and HTH associated domain; IPR007630, RNA polymerase sigma-70 region 4; IPR029291, DNA binding protein Tfx, C-terminal; IPR001789, Signal transduction response regulator, receiver domain; IPR013249, RNA polymerase sigma factor 70, region 4 type 2; IPR000014, PAS domain_].
FIGURE 3
Domain diagram of 110 LuxR containing Archaeal Proteins.
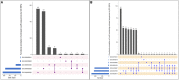
FIGURE 4
UpSetR plot showing distribution of Gene Ontology annotating function for LuxR containing proteins of Archaea in (A) Biological processes, and (B) Molecular functions. [_“regulation of transcription, DNA-templated” [GO:0006355]; “DNA-templated transcription, initiation” [GO:0006352]; “phosphorelay signal transduction system” [GO:0000160]; “transcription, DNA-templated” [GO:0006351]; “developmental process” [GO:0032502]; “DNA binding” [GO:0003677], “Endonuclease activity” [GO:0004519]; “Kinase activity” [GO:0016301]; “Sequence-specific DNA binding” [GO:0043565]; “Phosphorelay sensor kinase activity” [GO:0000155]; “Sigma factor activity” [GO:0016987]; “Transcription factor activity, sequence-specific DNA binding” [GO:0003700]_].
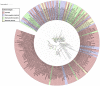
FIGURE 5
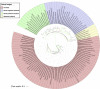
Phylogenetic analyses of 110 LuxR containing archaeal sequences along with 76 BLAST hits (bacteria and Archaea) using Maximum likelihood tree with bootstrap value of 1000.
FIGURE 6
Phylogenetic analyses of 16s rRNA sequences of 65 archaea along with 42 bacteria using Maximum likelihood tree with bootstrap value of 1000.
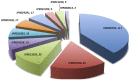
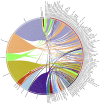
FIGURE 7
CIRCOS plot for representing the relationship between taxonomy and ecological niche. Circle in divided in two parts, rightmost showing 94 unique Archaeal strains and leftmost showing 27 unique habitats. Color of the right arc depicting 05 different groups (white, DPANN; gray, environmental samples; red, Euryarchaeota; black, TACK; light blue, unclassified Archaea) and left arch and the rays (links) are divided in 27 different colors (gray, Sulfur-reducing; light red, Organotrophic; green, Facultative organotrophic; pale red, Organoheterotrophic; very light blue, Chemoorganotrophic; blue, Carboxydotrophic; pale blue, Ammonia-oxidizing; orange, Hydrogenotrophic; pale purple, Hydrogen-producing; very very dark red, Nitrite-reducing; very very dark pale red, Nitrate-reducing; very very dark green, Aerobic; very very dark pale green, Facultatively aerobic; very very dark blue, Obligate anaerobic; very very dark pale blue, Strictly anaerobic; very very dark purple, Anaerobic; very very dark pale purple, Heterotrophic; very light dark grey, Methylotrophic; light blue, Methanogen; red, Neutrophile; very very dark pale orange, Mesophilic; very dark pale red, Barophilic; very very dark yellow, Hyperthermophilic; light pale green, Thermophilic; light orange, Extreme halophilic; light purple, Halophilic; black, Extreme haloalkaliphilic). Links of 27 colors showing the starting from habitat arc and ending in archaea arc showing their correlation.
Similar articles
- Bacterial LuxR solos have evolved to respond to different molecules including signals from plants.
Patel HK, Suárez-Moreno ZR, Degrassi G, Subramoni S, González JF, Venturi V. Patel HK, et al. Front Plant Sci. 2013 Nov 12;4:447. doi: 10.3389/fpls.2013.00447. eCollection 2013. Front Plant Sci. 2013. PMID: 24273546 Free PMC article. Review. - LuxR solos in Photorhabdus species.
Brameyer S, Kresovic D, Bode HB, Heermann R. Brameyer S, et al. Front Cell Infect Microbiol. 2014 Nov 18;4:166. doi: 10.3389/fcimb.2014.00166. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25478328 Free PMC article. - A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria.
Subramoni S, Florez Salcedo DV, Suarez-Moreno ZR. Subramoni S, et al. Front Cell Infect Microbiol. 2015 Feb 24;5:16. doi: 10.3389/fcimb.2015.00016. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25759807 Free PMC article. - LuxR Solos from Environmental Fluorescent Pseudomonads.
Bez C, Covaceuszach S, Bertani I, Choudhary KS, Venturi V. Bez C, et al. mSphere. 2021 Mar 31;6(2):e01322-20. doi: 10.1128/mSphere.01322-20. mSphere. 2021. PMID: 33789944 Free PMC article. - [Advances in quorum-sensing LuxR solos in bacteria].
Ye X, Zhu J, Wang H. Ye X, et al. Wei Sheng Wu Xue Bao. 2017 Mar 4;57(3):341-9. Wei Sheng Wu Xue Bao. 2017. PMID: 29756433 Review. Chinese.
Cited by
- Computer-Aided Rational Engineering of Signal Sensitivity of Quorum Sensing Protein LuxR in a Whole-Cell Biosensor.
Li J, Liu R, Chen Y, Liu S, Chen C, Liu T, Yang S, Zhuang Y, Yang R, Cui Y, Song Y, Wang T, Teng Y. Li J, et al. Front Mol Biosci. 2021 Aug 13;8:729350. doi: 10.3389/fmolb.2021.729350. eCollection 2021. Front Mol Biosci. 2021. PMID: 34485387 Free PMC article. - In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria.
Rajput A, Kumar M. Rajput A, et al. Sci Rep. 2017 Aug 1;7(1):6969. doi: 10.1038/s41598-017-07241-5. Sci Rep. 2017. PMID: 28765541 Free PMC article. - Synthetic Homoserine Lactone Sensors for Gram-Positive Bacillus subtilis Using LuxR-Type Regulators.
Zeng M, Sarker B, Howitz N, Shah I, Andrews LB. Zeng M, et al. ACS Synth Biol. 2024 Jan 19;13(1):282-299. doi: 10.1021/acssynbio.3c00504. Epub 2023 Dec 11. ACS Synth Biol. 2024. PMID: 38079538 Free PMC article. - Computational tools for exploring peptide-membrane interactions in gram-positive bacteria.
Kumar S, Balaya RDA, Kanekar S, Raju R, Prasad TSK, Kandasamy RK. Kumar S, et al. Comput Struct Biotechnol J. 2023 Mar 2;21:1995-2008. doi: 10.1016/j.csbj.2023.02.051. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36950221 Free PMC article. Review. - aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance.
Rajput A, Thakur A, Sharma S, Kumar M. Rajput A, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D894-D900. doi: 10.1093/nar/gkx1157. Nucleic Acids Res. 2018. PMID: 29156005 Free PMC article.
References
- Bala A., Gupta R. K., Chhibber S., Harjai K. (2013). Detection and quantification of quinolone signalling molecule: a third quorum sensing molecule of Pseudomonas aeruginosa by high performance-thin layer chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 930 30–35. 10.1016/j.jchromb.2013.04.027 - DOI - PubMed
- Borrel G., Harris H. M., Parisot N., Gaci N., Tottey W., Mihajlovski A., et al. (2013). Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc. 1:e00453–13. 10.1128/genomeA.00453-13 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources