Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases - PubMed (original) (raw)
Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases
Kuntal Pal et al. Structure. 2017.
Abstract
The nuclear pore complex subunit TPR is found in at least five different oncogenic fusion kinases, including TPR-MET, yet how TPR fusions promote activation of kinases and their oncogenic activities remains poorly understood. Here we report the crystal structure of TPR(2-142), the MET fusion partner of oncogenic TPR-MET. TPR(2-142) contains a continuous 124-residue α helix that forms an antiparallel tetramer from two leucine zipper-containing parallel coiled coils. Remarkably, single mutations cause strikingly different conformations of the coiled coil, indicating its highly dynamic nature. We further show that fusion of TPR(2-142) to the MET intracellular domain strongly and selectively stabilizes the αG helix of the MET kinase domain, and mutations of only the TPR leucine zipper residues at the junction to MET, but not other leucine zipper residues, abolish kinase activation. Together, these results provide critical insight into the TPR structure and its ability to induce dimerization and activation of fusion kinases.
Keywords: MET; oncogenic fusion kinase; receptor dimerization; receptor tyrosine kinase.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
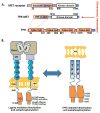
Figure 1. Schematic representation of the TPR-MET chromosomal translocation
**(A)**Structural and functional domains of MET receptor, Translocated promoter region (TPR), and the TPR(1-142)-MET(1010-1390) fusion protein. Chromosomal translocation mediated fusion between the Translocated Promoter Region (TPR) and the MET receptor genes are indicated by blue lines. Translocation results in expression of a cytosolic, constitutively active TPR-MET oncogene. (B) MET consists of a large extracellular region that contains a Sema domain, a cysteine-rich (CR) domain, and four immunoglobulin (IG) domains followed by a single transmembrane helix and the intracellular juxtamembrane and kinase domains. Hepatocyte growth factor (HGF)-regulated MET dimerization in the extracellular region transmits a signal for kinase domain (KD) autophosphorylation and downstream signaling. TPR-MET lacks the MET regulatory extracellular domain and forms a dimeric, constitutively active fusion kinase oncogene.
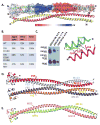
Figure 2. Structural organization of dimeric TPR (See also Figures S1 and S2)
(A) Crystal structure of TPR(2-142). TPR forms a parallel coiled coil dimer with chain A colored in green and chain B in cyan that further oligomerizes to form antiparallel tetramers (red and yellow chains). (B) Ribbon diagram of the helical caps of Chain A (green) and Chain B (cyan) with hydrophobic and ionic interactions indicated by dashed lines. (C–F) Stick representations of dimerization interface residues in the N-terminal coiled coil, the first leucine zipper (residues 77-100, D), the second leucine zipper (residues 120-141, E), and the middle region with a C75-C75 disulfide bond found in one of the tetramers in the asymmetric unit. The side chains of ‘a’ and ‘d’ residues in heptad (a-b-c-d-e-f-g) repeats of leucine zippers are displayed. (G) Disulfide crosslinking validation of the parallel arrangement of the TPR dimer. Cells transfected with TPR mutant constructs were treated with H2O2 and lysed, lysates separated by non-reducing SDS PAGE, and TPR and TPR crosslinking adducts detected by immunoblotting.
Figure 3. Tetrameric organization of TPR (See also Figures S3)
(A) TPR(2-142) tetramer in ribbon presentation, in which one dimer is overlaid with a transparent surface electrostatic potential map. Blue colored regions indicate positive charge and red colored regions negative charge (see charge potential heat map at bar below). The four monomers are colored green, cyan, magenta and yellow. (B) Tetramer distribution of TPR(2-142) wildtype and TPR-F55M/F113M(2-142) determined by size exclusion chromatography. (C) Validation of the antiparallel tetramer arrangement by in cell disulfide crosslinking between indicated residues of chain A and chain C. Lysates of H2O2-treated FLAG-TPR-MET expressing cells were analyzed by non-reducing SDS PAGE and immunoblotting with anti-FLAG antibody. (D) Parallel tetramer orientation of TPR(2-142) F113M. (E) Dimeric arrangement of TPR(2-142) F55M/F113M.
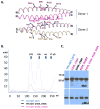
Figure 4. The TPR dimer-dimer interface is mediated largely by ionic interactions
(A.) Cartoon structure of the dimer-dimer interface between chain A (purple) and chain C (magenta). Key interface residues are shown in stick presentation. (B). Size column profiles of TPR-MET WT and TPR mutant proteins with charge reversal mutations of tetramer interface residues. Elution of size standards is indicated by arrows. The molecular weight of the monomeric TPR-MET fusion protein is 55.6 kDa, indicating that the peaks correspond to aggregated (~122 ml elution volume) and predominantly tetrameric (~169 ml) TPR-MET. (C). Immunoblot of lysates from AD293 cells expressing TPR-MET wildtype and tetramer interface mutants. TPR activity of mutant proteins was probed with antibodies that recognize the TPR kinase domain phosphorylated at Y1234 and Y1235 (pMET).
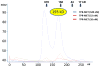
Figure 5. TPR-MET forms a stable tetramer
Size exclusion chromatogram of TPR-MET at different concentrations. Blue line: chromatogram of TPR-MET Ni eluate, which resolved two major peaks and allowed us to isolate the predominantly tetramer peak at ~169 ml elution volume and a concentration of 520 nM protein. We confirmed tetramer formation in the 169 ml eluate by MALS (calculated molecular weight: 193 kDa), and reloaded at lower concentrations onto the size exclusion column, resulting in only slightly right-shifted peaks at concentrations of 52 nM (brown line) and 26 nM (red line), indicating stable tetramer formation.
Figure 6. Hydrogen deuterium exchange mass spectrometry (HDX) analysis of monomeric and tetrameric MET kinase domains (See also Figures S4)
(A) MET kinase HDX perturbation map. Heat map of the difference in hydrogen deuterium exchange between the monomeric MET kinase domain and the tetrameric domain in the context of TPR(2-142)-MET overlaid on the MET kinase domain structure (PDB: 3Q6U). (B) Structure overlay of MET kinase domain in active (PDB: 3Q6U, green), inhibitor-bound inactive (PDB: 2WD1, orange), and non-phosphorylated inactive (PDB: 2G15, grey) conformations.
Figure 7. Mutational analysis of the TPR leucine zipper and the MET juxtamembrane domain (See also Figures S5)
(A) Leucine residues of the two TPR leucine zippers were mutated individually or in combination. Wildtype and mutant total (MET) and active, phosphorylated (pMET) TPR-MET were detected by immunoblotting. (B) Autophosphorylation of TPR-MET variants with truncations in the MET juxtamembrane domain. Left: Schematic of constructs analyzed. Right: Immunoblot probed with antibodies for total (MET) and active, phosphorylated (pMET) TPR-MET.
Similar articles
- Dimerization mediated through a leucine zipper activates the oncogenic potential of the met receptor tyrosine kinase.
Rodrigues GA, Park M. Rodrigues GA, et al. Mol Cell Biol. 1993 Nov;13(11):6711-22. doi: 10.1128/mcb.13.11.6711-6722.1993. Mol Cell Biol. 1993. PMID: 8413267 Free PMC article. - Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET.
Vigna E, Gramaglia D, Longati P, Bardelli A, Comoglio PM. Vigna E, et al. Oncogene. 1999 Jul 22;18(29):4275-81. doi: 10.1038/sj.onc.1202791. Oncogene. 1999. PMID: 10435641 - Oncogenic activation of the Met receptor tyrosine kinase fusion protein, Tpr-Met, involves exclusion from the endocytic degradative pathway.
Mak HH, Peschard P, Lin T, Naujokas MA, Zuo D, Park M. Mak HH, et al. Oncogene. 2007 Nov 8;26(51):7213-21. doi: 10.1038/sj.onc.1210522. Epub 2007 May 28. Oncogene. 2007. PMID: 17533376 - Pharmacological interference with protein-protein interactions mediated by coiled-coil motifs.
Strauss HM, Keller S. Strauss HM, et al. Handb Exp Pharmacol. 2008;(186):461-82. doi: 10.1007/978-3-540-72843-6_19. Handb Exp Pharmacol. 2008. PMID: 18491064 Review. - Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1.
Niemann HH. Niemann HH. Biochim Biophys Acta. 2013 Oct;1834(10):2195-204. doi: 10.1016/j.bbapap.2012.10.012. Epub 2012 Oct 31. Biochim Biophys Acta. 2013. PMID: 23123275 Review.
Cited by
- Overcoming resistance to targeted therapy using MET inhibitors in solid cancers: evidence from preclinical and clinical studies.
Ayoub NM, Ibrahim DR, Alkhalifa AE. Ayoub NM, et al. Med Oncol. 2021 Oct 19;38(12):143. doi: 10.1007/s12032-021-01596-6. Med Oncol. 2021. PMID: 34665336 Review. - Kinase regulation by liquid-liquid phase separation.
López-Palacios TP, Andersen JL. López-Palacios TP, et al. Trends Cell Biol. 2023 Aug;33(8):649-666. doi: 10.1016/j.tcb.2022.11.009. Epub 2022 Dec 15. Trends Cell Biol. 2023. PMID: 36528418 Free PMC article. Review. - B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Li Y, Ma X, Chen Z, Wu H, Wang P, Wu W, Cheng N, Zeng L, Zhang H, Cai X, Chen SJ, Chen Z, Meng G. Li Y, et al. Nat Commun. 2019 Aug 22;10(1):3789. doi: 10.1038/s41467-019-11746-0. Nat Commun. 2019. PMID: 31439836 Free PMC article. - Conserved regulatory motifs in the juxtamembrane domain and kinase N-lobe revealed through deep mutational scanning of the MET receptor tyrosine kinase domain.
Estevam GO, Linossi EM, Macdonald CB, Espinoza CA, Michaud JM, Coyote-Maestas W, Collisson EA, Jura N, Fraser JS. Estevam GO, et al. bioRxiv [Preprint]. 2024 May 6:2023.08.03.551866. doi: 10.1101/2023.08.03.551866. bioRxiv. 2024. PMID: 37577651 Free PMC article. Updated. Preprint. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
References
- Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nature reviews Molecular cell biology. 2017;18:73–89. - PubMed
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nature reviews Molecular cell biology. 2003;4:915–925. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous