PAR-1 promotes microtubule breakdown during dendrite pruning in Drosophila - PubMed (original) (raw)
PAR-1 promotes microtubule breakdown during dendrite pruning in Drosophila
Svende Herzmann et al. EMBO J. 2017.
Abstract
Pruning of unspecific neurites is an important mechanism during neuronal morphogenesis. Drosophila sensory neurons prune their dendrites during metamorphosis. Pruning dendrites are severed in their proximal regions. Prior to severing, dendritic microtubules undergo local disassembly, and dendrites thin extensively through local endocytosis. Microtubule disassembly requires a katanin homologue, but the signals initiating microtubule breakdown are not known. Here, we show that the kinase PAR-1 is required for pruning and dendritic microtubule breakdown. Our data show that neurons lacking PAR-1 fail to break down dendritic microtubules, and PAR-1 is required for an increase in neuronal microtubule dynamics at the onset of metamorphosis. Mammalian PAR-1 is a known Tau kinase, and genetic interactions suggest that PAR-1 promotes microtubule breakdown largely via inhibition of Tau also in Drosophila Finally, PAR-1 is also required for dendritic thinning, suggesting that microtubule breakdown might precede ensuing plasma membrane alterations. Our results shed light on the signaling cascades and epistatic relationships involved in neurite destabilization during dendrite pruning.
Keywords: PAR‐1; Tau; dendrite; pruning.
© 2017 The Authors.
Figures
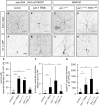
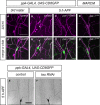
Figure 1. PAR‐1 is required for sensory neuron dendrite pruning
- A–D′
Loss of PAR‐1 causes defects in c4da neuron dendrite pruning. Upper panels (A–D) show third‐instar larval neurons, and lower panels (A′–D′) show neurons at 18 h APF. (A, A′) Control c4da neurons labeled by UAS‐CD8GFP expression under the control of ppk‐GAL4 (third chromosome insertion). (B, B′) C4da neurons expressing par‐1 RNAi under ppk‐GAL4. (C, C′) MARCM clones of par‐1 Δ16 mutant c4da neurons. (D, D′) Rescue of par‐1 Δ16 mutant MARCM c4da neuron pruning defects by UAS‐mediated expression of wild‐type PAR‐1 (isoform RR). - E
Percentages of neurons with dendrite pruning defects. ***P < 0.0005, *P < 0.05 (using Fisher's exact test). N = 16–38. - F
Number of attached primary and secondary dendrites at 18 h APF. ***P < 0.0005, **P < 0.005 (using Wilcoxon's test). N = 16–38. - G
Length of unpruned dendrites at 18 h APF. ***P < 0.0005 (using Wilcoxon's test). N = 16–38.
Data information: Scale bars are 100 μm in (A–D) and 50 μm in (A′–D′). Error bars represent s.d.
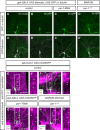
Figure 2. PAR‐1 is required for dendritic microtubule breakdown during the early phase of c4da neuron dendrite pruning
- A–D′
Microtubules were labeled by expression of UAS‐GFP::α‐tubulin in c4da neurons under ppk‐GAL4, and c4da neuron morphology was visualized by UAS‐tdtomato. Panels (A–D) show GFP::α‐tubulin staining, and panels (A′–D′) show the merge with tdtomato. (A, A′) Third‐instar control c4da neuron. The asterisk denotes a c3da neuron that is also sometimes labeled by the ppk‐GAL4 driver. (B, B′) Control c4da neuron at 5 h APF. GFP signal disappears from proximal dendrite regions. (C, C′) C4da neuron expressing par‐1 RNAi at 5 h APF. (D, D′) par‐1 Δ16 mutant c4da neuron MARCM clone at 5 h APF. Continuous GFP staining persists in proximal dendrites after loss of PAR‐1. - E–H′
Microtubules were labeled by an antibody against acetylated α‐tubulin, and c4da neuron morphology was visualized by UAS‐CD8GFP expressed under ppk‐GAL4, or by tdtomato in MARCM clones. Panels (E–H) show merges of the indicated genotypes, and panels (E′–H′) show only the acetylated α‐tubulin signal of the boxed regions in (E–H). Arrows indicate the positions of dendrites. (E, E′) Third‐instar control c4da neuron. (F, F′) Control c4da neuron at 5 h APF. (G, G′) C4da neuron expressing par‐1 RNAi at 5 h APF. (H, H′) par‐1 Δ16 mutant c4da neuron MARCM clone at 5 h APF.
Data information: Scale bars are 50 μm.
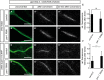
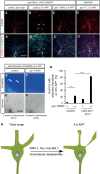
Figure 3. PAR‐1 is required for increased microtubule dynamics in c4da neurons at the onset of the pupal phase
Photoconvertible tdEOS::α‐tubulin was expressed in c4da neurons under ppk‐GAL4. tdEOS::α‐tubulin was photoconverted in small dendrite segments with a 405‐nm laser at the indicated developmental stages, and decay of converted material was assessed after 30 min. Panels (A, B, D, E) show unconverted green tdEOS::α‐tubulin signal to demarcate dendrites, panels (A′, B′, D′, E′) show converted red tdEOS::α‐tubulin immediately after conversion, and panels (A″, B″, D″, E″) show the converted material 30 min after conversion.
- A–A″
Third‐instar control c4da neuron. - B–B″
Third‐instar c4da neuron expressing par‐1 RNAi. - C
Quantification of remaining red tdEOS::α‐tubulin in panels (A and B). N was 18 (control) and 15 (par‐1 RNAi), respectively. P = 0.098, Wilcoxon's test. - D–D″
White pupal control c4da neuron (0 h APF). - E–E″
White pupal c4da neuron expressing par‐1 RNAi (0 h APF). - F
Quantification of remaining red tdEOS in panels (D and E). N was 34 (control) and 35 (par‐1 RNAi). ***P < 0.0005, Wilcoxon's test.
Data information: Scale bars are 5 μm. Error bars represent s.d.
Figure EV1. PAR‐1 phosphorylates Drosophila tau
- The indicated combinations of recombinant FLAGPAR‐1 (isoform RR, wild‐type, or kinase‐dead K510A) from S2 cells and bacterially expressed GST‐tau were incubated with ATPγS, and thiophosphorylated proteins were detected after alkylation with an antibody against a semisynthetic thiophosphate epitope (anti‐hapten) (Allen et al, 2007).
- Phosphoassay with wild‐type FLAGPAR‐1 and the indicated GST‐tau phosphomutants.
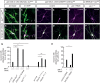
Figure 4. Tau distribution in da neurons and genetic interactions with PAR‐1 during dendrite pruning
- A–B′
Endogenous Tau was visualized using a MiMIC‐derived Tau::GFP fusion protein. Arrows mark c4da neuron dendrites. (A, A′) Endogenous Tau localizes to axons and dendrites in third‐instar c4da neurons. (B, B′) Tau is reduced in dendrites at 5 h APF. - C–F′
Tau distribution in c4da neurons using transgenic HA‐tagged Tau. Neurons were labeled by UAS‐CD8GFP under ppk‐GAL4 (C–E) or by UAS‐tdtomato in MARCM clones (F). (C, C′) TauHA staining in a third‐instar control neuron. (D, D′) TauHA staining in a control neuron at 5 h APF. (E, E′) TauHA distribution in a c4da neuron expressing par‐1 RNAi at 5 h APF. (F, F′) TauHA distribution in par‐1 Δ16 mutant c4da neuron at 5 h APF. - G
Specific dosage‐dependent genetic interactions between par‐1 and tau. Effects of Tau or Futsch upregulation (UAS‐TauHA, UAS‐TauHA S184A, futsch EP1419) or downregulation (tau MR22/+, futsch K68 /Y) on dendrite pruning defects induced by par‐1 RNAi at 18 h APF. All transgenes were expressed under the control of a second chromosome insertion of ppk‐GAL4. ***P < 0.0005, *P < 0.05, Fisher's exact test, N = 23–48. - H
Genetic interactions between par‐1 and kat‐60L1. par‐1 RNAi was coexpressed with Or83b RNAi as a control, or with kat‐60L1 RNAi, and the effects on dendrite pruning were assessed at 18 h APF. **P < 0.005, Fisher's exact test. N = 38–49.
Data information: Scale bars are 30 μm in (A–B′), and 25 μm in (C–F′).
Figure EV2. Futsch/MAP1B distribution during c4da neuron dendrite pruning and effects of tau depletion on dendrite pruning
- A–D′
Futsch/22C10 distribution in c4da neurons. Panels (A–D) show Futsch/22C10 staining, and panels (A′–D′) show the merge with the c4da neuron marker (ppk>CD8GFP in A–C, tdtomato in D). (A, A′) Control c4da neuron at the third‐instar larval stage. (B, B′) Control c4da neuron at 5 h APF. Futsch/22C10 is lost from proximal dendrites (arrows). (C, C′) A c4da neuron expressing par‐1 RNAi at 5 h APF still shows strong dendritic Futsch/22C10. (D, D′) A par‐1 Δ_16_ mutant MARCM c4da neuron at 5 h APF still shows strong dendritic Futsch/22C10. - E, F
tau RNAi does not trigger precocious dendrite pruning at 5 h APF. Representative images of control c4da neurons (E) or c4da neurons expressing tau RNAi under the control of ppk‐GAL4 (F) are shown.
Data information: Scale bars are 50 μm.
Figure 5. PAR‐1 is required for c1da neuron dendrite pruning in a _tau_‐sensitive manner
Dorsal c1da neurons (ddaD) of the indicated genotypes were labeled by GAL4 2–21 driving expression of UAS‐tdtomato (A, B) or by MARCM (C–F) and imaged at third instar or at 18 h APF.
- Third‐instar larval c1da neurons. The c1da neuron ddaD is marked by an arrow.
- At 18 h APF, c1da neurons have largely pruned their larval dendrites. Arrow, ddaD.
- A par‐1 Δ16 mutant c1da neuron retains its dendrites at 18 h APF.
- A par‐1 Δ16 mutant c1da neuron expressing wild‐type PAR‐1 (isoform RR).
- A par‐1 Δ16 mutant c1da neuron in a heterozygous tau MR22 /+ mutant background.
- A par‐1 Δ16 mutant c1da neuron expressing wild‐type PAR‐1 (isoform RL).
- A par‐1 Δ16 mutant c1da neuron expressing kinase‐dead PAR‐1 (isoform RL).
- Quantification of numbers of attached primary and secondary dendrites at 18 h APF (N = 13–25 for the MARCM experiments). ***P < 0.0005, *_P_ < 0.05. n. s., not significant, _P_ > 0.05 (using Wilcoxon's test). Error bars represent s.d.
Data information: Scale bars are 50 μm.
Figure 6. PAR‐1 is required for dendritic membrane alterations during pruning
- A–C′
Ank2XL is lost from proximal dendrites in a PAR‐1‐dependent manner during the early pupal stage. Upper panels (A–C) show Ank2XL staining at the indicated developmental stages, and lower panels (A′–C′) show merge with Futsch/22C10 staining and c4da neuron markers. (A, A′) Third‐instar larval control c4da neuron. (B, B′) Control c4da neuron at 5 h APF. Arrows indicate dendrite regions devoid of Ank2XL and 22C10 staining. (C, C′) C4da neuron expressing par‐1 RNAi at 5 h APF. - D, D′
par‐1 Δ16 mutant c4da neuron at 5 h APF. - E–F′
PAR‐1 is required for dendritic Ca2+ transients during the early phase of pruning. Transgenes were expressed under ppk‐GAL4. Panels (E and F) show GCaMP6s fluorescence intensity in c4da neuron dendrites at 6 h APF, and panels (E′ and F′) show the tdtomato marker to visualize neuronal morphology. Numbers below panels indicate the average number of dendrites with independent Ca2+ transients (branch units) in a 10‐min movie (N = 6). (E, E′) Control c4da neuron. Arrows indicate dendrites with Ca2+ transients. (F, F′) C4da neuron expressing par‐1 RNAi. - G
Genetic interactions between PAR‐1 and shibire/dynamin. Dendrite pruning defects of the indicated genotypes were analyzed at 18 h APF as in Fig 4. All flies were kept at 25°C, the permissive temperature for shi ts. ***P < 0.0005, **P < 0.005, Fisher's exact test. N = 48–64. - H
Model. Activation of PAR‐1 leads to loss of c4da neuron dendritic microtubules via Tau inhibition and possibly, Kat‐60L1 activation. Microtubule loss in proximal regions precedes dendritic thinning.
Data information: Scale bars are 50 μm.
Similar articles
- Spatial regulation of microtubule disruption during dendrite pruning in Drosophila.
Herzmann S, Götzelmann I, Reekers LF, Rumpf S. Herzmann S, et al. Development. 2018 May 11;145(9):dev156950. doi: 10.1242/dev.156950. Development. 2018. PMID: 29712642 - Drosophila CLASP regulates microtubule orientation and dendrite pruning by suppressing Par-1 kinase.
Bu S, Tang Q, Wang Y, Lau SSY, Yong WL, Yu F. Bu S, et al. Cell Rep. 2022 May 31;39(9):110887. doi: 10.1016/j.celrep.2022.110887. Cell Rep. 2022. PMID: 35649352 - Drosophila GSK3β promotes microtubule disassembly and dendrite pruning in sensory neurons.
Dzaki N, Bu S, Lau SSY, Yong WL, Yu F. Dzaki N, et al. Development. 2022 Nov 15;149(22):dev200844. doi: 10.1242/dev.200844. Epub 2022 Nov 16. Development. 2022. PMID: 36264221 - Functions of Microtubule Disassembly during Neurite Pruning.
Rumpf S, Wolterhoff N, Herzmann S. Rumpf S, et al. Trends Cell Biol. 2019 Apr;29(4):291-297. doi: 10.1016/j.tcb.2019.01.002. Epub 2019 Jan 22. Trends Cell Biol. 2019. PMID: 30683460 Review. - Spatiotemporal regulation of developmental neurite pruning: Molecular and cellular insights from Drosophila models.
Furusawa K, Emoto K. Furusawa K, et al. Neurosci Res. 2021 Jun;167:54-63. doi: 10.1016/j.neures.2020.11.010. Epub 2020 Dec 10. Neurosci Res. 2021. PMID: 33309868 Review.
Cited by
- A microtubule polymerase is required for microtubule orientation and dendrite pruning in Drosophila.
Tang Q, Rui M, Bu S, Wang Y, Chew LY, Yu F. Tang Q, et al. EMBO J. 2020 May 18;39(10):e103549. doi: 10.15252/embj.2019103549. Epub 2020 Apr 8. EMBO J. 2020. PMID: 32267553 Free PMC article. - Microtubule dynamics in healthy and injured neurons.
Rolls MM, Thyagarajan P, Feng C. Rolls MM, et al. Dev Neurobiol. 2021 Apr;81(3):321-332. doi: 10.1002/dneu.22746. Epub 2020 Apr 25. Dev Neurobiol. 2021. PMID: 32291942 Free PMC article. Review. - Spatiotemporal changes in microtubule dynamics during dendritic morphogenesis.
Hu C, Feng P, Chen M, Tang Y, Soba P. Hu C, et al. Fly (Austin). 2022 Dec;16(1):13-23. doi: 10.1080/19336934.2021.1976033. Fly (Austin). 2022. PMID: 34609266 Free PMC article. - JNK signaling coordinates with ecdysone signaling to promote pruning of Drosophila sensory neuron dendrites.
Zhu S, Chen R, Soba P, Jan YN. Zhu S, et al. Development. 2019 Apr 17;146(8):dev163592. doi: 10.1242/dev.163592. Development. 2019. PMID: 30936183 Free PMC article. - Establishment of a novel axon pruning model of Drosophila motor neuron.
Xu W, Kong W, Gao Z, Huang E, Xie W, Wang S, Rui M. Xu W, et al. Biol Open. 2023 Jan 1;12(1):bio059535. doi: 10.1242/bio.059535. Epub 2023 Jan 6. Biol Open. 2023. PMID: 36606515 Free PMC article.
References
- Benton R, St Johnston D (2003) Drosophila PAR‐1 and 14‐3‐3 inhibit Bazooka/PAR‐3 to establish complementary cortical domains in polarized cells. Cell 115: 691–704 - PubMed
- Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E (1993) Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF‐like immunoreactivity and microtubule binding. Neuron 11: 153–163 - PubMed
- Brill MS, Kleele T, Ruschkies L, Wang M, Marahori NA, Reuter MS, Hausrat TJ, Weigand E, Fisher M, Ahles A, Engelhardt S, Bishop DL, Kneussel M, Misgeld T (2016) Branch‐specific microtubule destabilization mediates axon branch loss during neuromuscular synapse elimination. Neuron 92: 845–856 - PMC - PubMed
- Cox DN, Lu B, Sun TQ, Williams LT, Jan YN (2001) Drosophila par‐1 is required for oocyte differentiation and microtubule organization. Curr Biol 11: 75–87 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials