Polyvalent Proteins, a Pervasive Theme in the Intergenomic Biological Conflicts of Bacteriophages and Conjugative Elements - PubMed (original) (raw)
. 2017 Jul 11;199(15):e00245-17.
doi: 10.1128/JB.00245-17. Print 2017 Aug 1.
Affiliations
- PMID: 28559295
- PMCID: PMC5512222
- DOI: 10.1128/JB.00245-17
Polyvalent Proteins, a Pervasive Theme in the Intergenomic Biological Conflicts of Bacteriophages and Conjugative Elements
Lakshminarayan M Iyer et al. J Bacteriol. 2017.
Abstract
Intense biological conflicts between prokaryotic genomes and their genomic parasites have resulted in an arms race in terms of the molecular "weaponry" deployed on both sides. Using a recursive computational approach, we uncovered a remarkable class of multidomain proteins with 2 to 15 domains in the same polypeptide deployed by viruses and plasmids in such conflicts. Domain architectures and genomic contexts indicate that they are part of a widespread conflict strategy involving proteins injected into the host cell along with parasite DNA during the earliest phase of infection. Their unique feature is the combination of domains with highly disparate biochemical activities in the same polypeptide; accordingly, we term them polyvalent proteins. Of the 131 domains in polyvalent proteins, a large fraction are enzymatic domains predicted to modify proteins, target nucleic acids, alter nucleotide signaling/metabolism, and attack peptidoglycan or cytoskeletal components. They further contain nucleic acid-binding domains, virion structural domains, and 40 novel uncharacterized domains. Analysis of their architectural network reveals both pervasive common themes and specialized strategies for conjugative elements and plasmids or (pro)phages. The themes include likely processing of multidomain polypeptides by zincin-like metallopeptidases and mechanisms to counter restriction or CRISPR/Cas systems and jump-start transcription or replication. DNA-binding domains acquired by eukaryotes from such systems have been reused in XPC/RAD4-dependent DNA repair and mitochondrial genome replication in kinetoplastids. Characterization of the novel domains discovered here, such as RNases and peptidases, are likely to aid in the development of new reagents and elucidation of the spread of antibiotic resistance.IMPORTANCE This is the first report of the widespread presence of large proteins, termed polyvalent proteins, predicted to be transmitted by genomic parasites such as conjugative elements, plasmids, and phages during the initial phase of infection along with their DNA. They are typified by the presence of multiple domains with disparate activities combined in the same protein. While some of these domains are predicted to assist the invasive element in replication, transcription, or protection of their DNA, several are likely to target various host defense systems or modify the host to favor the parasite's life cycle. Notably, DNA-binding domains from these systems have been transferred to eukaryotes, where they have been incorporated into DNA repair and mitochondrial genome replication systems.
Keywords: DNA replication; DNA-binding proteins; RNases; antirestriction; bacteriophages; biological conflicts; effectors; metallopeptidase; plasmids; transcription.
Copyright © 2017 Iyer et al.
Figures
FIG 1
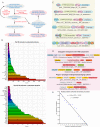
(A) Schematic showing the recursive process used to find domains in polyvalent proteins. (B) Frequency distribution of the top 100 domains observed in polyvalent proteins. (C) Examples of recovered polyvalent proteins and their domain architectures. (D) Examples of neighborhoods of polyvalent-protein-encoding genes illustrating the broad types of genome contexts. (E) Plot illustrating the number of polyvalent proteins with a given number of domains. Note that the y axis is on a log scale.
FIG 2
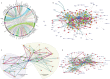
Polyvalent protein domain networks. (A) Chord diagram of domain cooccurrences in polyvalent proteins. The plot includes all cooccurrences of domains. Thus, an edge is drawn between two domains in a protein whether they are adjacent or not. (B) Domain architecture network of polyvalent proteins. Domains linked in the same protein are connected by arrows with each arrowhead pointing to the C-terminal domain. Here and in the subsequent network images, an edge is drawn only between two adjacent domains. (C) Clique subnetwork of the domain architecture network merging large cliques with seven or eight nodes. The network reveals two distinct subgraphs as described in the text. (D) Largest biconnected subnetwork of the domain architecture network. In all of the networks, the node size and color are scaled on the basis of the number of connections per node (degree). Nodes with two or fewer connections are gray. Edge thickness is based on the number of edge occurrences. Edges occurring <15 times are gray, those occurring 16 to 90 times are cadet blue, and those occurring >90 times are maroon.
FIG 3
Multiple-sequence alignments of MPTase (A), Pol-β NTase (B), DdrB-ParB (C), and PBECR (D) domains. Secondary structure and element labeling is provided at the top line, and residue consensus at various percentages is provided at the bottom line. Sequences are identified on the left by gene name, organism abbreviation, and NCBI accession number separated by vertical lines and to the right by family name. Sequences of structures are labeled with PDB codes, shaded in orange. Poorly conserved secondary structure elements are colored white. Family-specific conserved residues described in the text are denoted by blue boxes. Alignments are colored as follows: h (hydrophobic), l (aliphatic), and a (aromatic) are shaded yellow; p (polar), + (positively charged), − (negatively charged), and c (charged) are shaded blue; s (small) and t (tiny) are shaded green; b (big) is shaded gray; absolutely conserved residues are in white lettering and shaded in black. For organism abbreviations, see File S4 at
ftp://ftp.ncbi.nih.gov/pub/aravind/polyvalent/polyvalent.html
.
FIG 4
Domain architectures and gene neighborhoods of domains found in polyvalent proteins grouped by the presence of various principal domains or groups of domains, including peptidases (A), ARTs (B), kinases (C), GNATs (D), Pol-β NTases (E), helicases (F), DdrB-like ParB (G), RadC (H), SMS/RadA (I), and Toprim fold and Primpol primases (J). These include only a small sample of the entire diaspora of associations. For the complete set of domain architectures and operons, see
ftp://ftp.ncbi.nih.gov/pub/aravind/polyvalent/polyvalent.html
. Proteins and gene neighborhoods are shown with their species names and GenBank accession numbers. For gene neighborhoods, the accession number of the gene in dark pink is used. Gene names are shown only for well-studied proteins. Genes in neighborhoods are shown as boxed arrows, with the arrowhead pointing to the 3′ gene. Domains are not drawn to scale.
FIG 5
Domain architectures and gene neighborhoods of domains found in polyvalent proteins grouped by the presence of various principal domains or groups of domains, including PBECR (A); 2H (B); RelA/SpoT (C); inorganic pyrophosphatases (D); phosphoribosyltransferases (E); peptidoglycan and cytoskeleton-targeting domains (F); nucleic acid binding domains (G); and DarA, ArdA, and YodL domains (H). Domain and gene neighborhood designations are as in Fig. 4.
Similar articles
- CoCoNuTs are a diverse subclass of Type IV restriction systems predicted to target RNA.
Bell RT, Sahakyan H, Makarova KS, Wolf YI, Koonin EV. Bell RT, et al. Elife. 2024 May 13;13:RP94800. doi: 10.7554/eLife.94800. Elife. 2024. PMID: 38739430 Free PMC article. - Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements.
Makarova KS, Wolf YI, van der Oost J, Koonin EV. Makarova KS, et al. Biol Direct. 2009 Aug 25;4:29. doi: 10.1186/1745-6150-4-29. Biol Direct. 2009. PMID: 19706170 Free PMC article. - Role of CRISPR/cas system in the development of bacteriophage resistance.
Szczepankowska A. Szczepankowska A. Adv Virus Res. 2012;82:289-338. doi: 10.1016/B978-0-12-394621-8.00011-X. Adv Virus Res. 2012. PMID: 22420856 Review. - Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing.
Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Anantharaman V, et al. Biol Direct. 2013 Jun 15;8:15. doi: 10.1186/1745-6150-8-15. Biol Direct. 2013. PMID: 23768067 Free PMC article. - Friendly Fire: Biological Functions and Consequences of Chromosomal Targeting by CRISPR-Cas Systems.
Heussler GE, O'Toole GA. Heussler GE, et al. J Bacteriol. 2016 Apr 28;198(10):1481-6. doi: 10.1128/JB.00086-16. Print 2016 May 15. J Bacteriol. 2016. PMID: 26929301 Free PMC article. Review.
Cited by
- Genome sequencing and assessment of plant growth-promoting properties of a Serratia marcescens strain isolated from vermicompost.
Matteoli FP, Passarelli-Araujo H, Reis RJA, da Rocha LO, de Souza EM, Aravind L, Olivares FL, Venancio TM. Matteoli FP, et al. BMC Genomics. 2018 Oct 16;19(1):750. doi: 10.1186/s12864-018-5130-y. BMC Genomics. 2018. PMID: 30326830 Free PMC article. - Tailed Lytic Bacteriophages of Soft Rot Pectobacteriaceae.
Miroshnikov KA, Evseev PV, Lukianova AA, Ignatov AN. Miroshnikov KA, et al. Microorganisms. 2021 Aug 26;9(9):1819. doi: 10.3390/microorganisms9091819. Microorganisms. 2021. PMID: 34576713 Free PMC article. Review. - Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts.
M Iyer L, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. M Iyer L, et al. Viruses. 2021 Jan 5;13(1):63. doi: 10.3390/v13010063. Viruses. 2021. PMID: 33466489 Free PMC article. - In Silico Characterization of Uncharacterized Proteins From Multiple Strains of Clostridium Difficile.
Abbasi BA, Dharan A, Mishra A, Saraf D, Ahamad I, Suravajhala P, Valadi J. Abbasi BA, et al. Front Genet. 2022 Aug 11;13:878012. doi: 10.3389/fgene.2022.878012. eCollection 2022. Front Genet. 2022. PMID: 36035185 Free PMC article. - Pectobacterium versatile Bacteriophage Possum: A Complex Polysaccharide-Deacetylating Tail Fiber as a Tool for Host Recognition in Pectobacterial Schitoviridae.
Lukianova AA, Evseev PV, Shneider MM, Dvoryakova EA, Tokmakova AD, Shpirt AM, Kabilov MR, Obraztsova EA, Shashkov AS, Ignatov AN, Knirel YA, Dzhalilov FS, Miroshnikov KA. Lukianova AA, et al. Int J Mol Sci. 2022 Sep 20;23(19):11043. doi: 10.3390/ijms231911043. Int J Mol Sci. 2022. PMID: 36232343 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources