AACR Project GENIE: Powering Precision Medicine through an International Consortium - PubMed (original) (raw)
AACR Project GENIE: Powering Precision Medicine through an International Consortium
AACR Project GENIE Consortium. Cancer Discov. 2017 Aug.
Abstract
The AACR Project GENIE is an international data-sharing consortium focused on generating an evidence base for precision cancer medicine by integrating clinical-grade cancer genomic data with clinical outcome data for tens of thousands of cancer patients treated at multiple institutions worldwide. In conjunction with the first public data release from approximately 19,000 samples, we describe the goals, structure, and data standards of the consortium and report conclusions from high-level analysis of the initial phase of genomic data. We also provide examples of the clinical utility of GENIE data, such as an estimate of clinical actionability across multiple cancer types (>30%) and prediction of accrual rates to the NCI-MATCH trial that accurately reflect recently reported actual match rates. The GENIE database is expected to grow to >100,000 samples within 5 years and should serve as a powerful tool for precision cancer medicine.Significance: The AACR Project GENIE aims to catalyze sharing of integrated genomic and clinical datasets across multiple institutions worldwide, and thereby enable precision cancer medicine research, including the identification of novel therapeutic targets, design of biomarker-driven clinical trials, and identification of genomic determinants of response to therapy. Cancer Discov; 7(8); 818-31. ©2017 AACR. See related commentary by Litchfield et al., p. 796 This article is highlighted in the In This Issue feature, p. 783.
©2017 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conficts of Interest: F. André reports receiving commercial research grants from Astra-Zeneca, Lilly, Novartis, and Pfzer. M. Arnedos has received honoraria from the speakers bureaus of Novartis and AstraZeneca, and is a consultant/advisory board member for Puma. D.M. Hyman reports receiving commercial research grants from AstraZeneca, Loxo Oncology, and PUMA Biotechnology, and is a consultant/advisory board member for Atara Biotherapeutics, Chugai, and CytomX. M.A. Levy is an advisory board member of Personalis, Inc., and receives royalty distribution from GenomOncology. F. Meric-Bernstam reports receiving commercial research grants from Aileron, AstraZeneca, Bayer, Calithera, Curis, CytoMx, Debiopharma, Effective Pharma, Genentech, Jounce, Novartis, PUMA, Taiho, and Zymeworks, and is a consultant/advisory board member for Clearlight Diagnostics, Darwin Health, Dialecta, GRAIL, Infection Biosciences, and Pieris. G.B. Mills reports receiving commercial research grants from Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcome Technologies, Illumina, Karus, Komen Research Foundation, NanoString, and Takeda/Millennium Pharmaceuticals; has received honoraria from the speakers bureaus of Allostery, AstraZeneca, ImmunoMet, ISIS Pharmaceuticals, Lilly, MedImmune, Novartis, Pfzer, Symphogen, and Tarveda; has ownership interest (including patents) in Catena Pharmaceuticals, ImmunoMet, Myriad Genetics, PTV Ventures, and Spindletop Ventures; and is a consultant/advisory board member for Adventist Health, Allostery, AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMet, ISIS Pharmaceuticals, Lilly, MedImmune, Novartis, Precision Medicine, Provista Diagnostics, Signalchem Lifesciences, Symphogen, Takeda/Millennium Pharmaceuticals, Tarveda, and Tau Therapeutics. T.J. Pugh is a consultant/advisory board member for Dynacare. C.L. Sawyers is a consultant/advisory board member for Novartis. V.E. Velculescu has ownership interest (including patents) in Personal Genome Diagnostics and is a consultant/advisory board member for the same. No potential conflicts of interest were disclosed by the other authors.\One of the Editors-in-Chief is an author on this article. In keeping with the AACR's editorial policy, the peer review of this submission was managed by a senior member of Cancer Discovery's editorial team; a member of the AACR Publications Committee rendered the final decision concerning acceptability.
Figures
Figure 1
AACR Project GENIE at a glance. A, Variant calls and a limited clinical dataset from patients treated at each of the participating centers are sent to the Synapse platform, developed by Sage Bionetworks, where the data are harmonized and protected health information (PHI) removed in a secure Health Insurance Portability and Accountability Act (HIPAA)-compliant environment that provides data governance. Once harmonized, these data are viewed and analyzed in the cBioPortal for Cancer Genomics. Value is provided to both the data generators and the consortium by establishing 6-month periods of exclusivity to each prior to the data becoming available to the broader research community. B, Once data are available in the cBioPortal, clinical research projects are proposed and vetted by the project steering committee. Clinical teams are then assembled to define the clinical attributes required to answer the approved research question; these data are then manually curated from the relevant medical records and deposited in an electronic data capture system. The detailed clinical data are then transferred to Synapse where they are linked with the appropriate genomic and limited clinical data and are viewable and analyzable in the cBioPortal platform. Again, value is created by providing a period of at least 6 months' exclusivity to both the consortium and sponsors, where relevant. The primary data are made public at the time of publication.
Figure 2
Landscape overview of GENIE dataset. A, The degree of overlap at the gene level across the contributing centers' genomic assays is shown. A core set of 44 genes (listed in the inlay) is represented across all genomic assays in the GENIE dataset. The 2 additional genes listed in the bottom right of the inlay in gray are genes that were common to the smaller panels, not present in some of the previous versions of the larger panels but are present on the most recent version of all panels. B, Total sample counts by tumor type and contributing center. The contribution of samples for each tumor type across the institutions in shown within each bar of the lower stacked barplot. C, Mutations (all nonsilent substitutions and small insertions/deletions reported) per coding megabase (Mbs) sequenced for each sample, stratified by tumor type, and ordered by median mutation rate in those tumor types. The data are shown as empirical cumulative distributions (blue-shaded area) with individual samples shown as points colored black to red for low to high mutation burden, respectively. These data are limited to the 14,310 samples analyzed by the larger gene panels used at centers DFCI, MSK, and VICC.
Figure 3
Genomic alterations in non-small cell lung cancer, breast cancer, and colorectal cancer. A-C, The genomic alteration rate (including mutation, copy number, and rearrangement) aggregated to the gene level across the cohort for the top three most common tumor types is shown: non-small cell lung cancer, colorectal cancer, and breast cancer (A-C, respectively). Data for each center are shown as percentage of samples from that center with genomic alterations in a given gene. Directly adjacent to the main heat map is the proportional breakdown of the types of genomic alterations observed, and characterization of the mutation distribution observed in a given gene as oncogene and tumor suppressor, based on the normalized entropy (log2(N)-Σ_pi_log2(pi), where N is the number of unique mutations in a given gene and pi is the proportion of mutations accounted for by a given unique mutation of a given gene) in the mutation spectrum and the prevalence of truncating and frameshift mutations, respectively. These data are limited to the gene with either: (i) 15% genomic alteration rate in at least one center, (ii) 5% genomic alteration rate in at least three centers, and (iii) OncoKB level 1 or 2A evidence for the tumor types shown. The “nc” designation in the colorbar legend indicates no coverage.
Figure 4
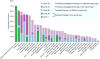
Potential clinical actionability. Tumor types are shown by decreasing overall frequency of actionability. Actionability was defined by the union of three knowledge bases: My Cancer Genome (
), OncoKB (
), and the Personalized Cancer Therapy knowledge base (
). For each tumor sample, the highest level of actionability of any variant was considered. Only tumor types with 100 or more samples were included in this analysis.
Figure 5
Clinical trial matching. Overview of GENIE samples matched to NCI-MATCH, based on genomic and cancer type criteria. Each patient with a reported sequencing date in 2014 or later was matched against 18 arms of the study that use somatic mutations or copy-number alterations for enrollment. Arms with fusion criteria were excluded because only two of the eight contributing GENIE centers provided fusion data. A, Information regarding 18 arms of the NCI-MATCH trial, including a summary of genomic trial eligibility, and the total count of GENIE samples matched. For arms S1 and U (indicated with an asterisk), the exact set of inactivating mutations was not specified in the NCI protocol, and all mutations were therefore considered matches. B, Proportion of the matches attributed to the top 10 most frequently matched cancer types. The categories are the top-level OncoTree codes. C, Comparison of the observed matching rate in the GENIE cohort with the reported rates observed by the first 645 patients by the NCI-MATCH group. Substudies X and Z1D had not reported interim rates.
Comment in
- The GENIE Is Out of the Bottle: Landmark Cancer Genomics Dataset Released.
Litchfield K, Turajlic S, Swanton C. Litchfield K, et al. Cancer Discov. 2017 Aug;7(8):796-798. doi: 10.1158/2159-8290.CD-17-0604. Cancer Discov. 2017. PMID: 28765114
Similar articles
- AACR Project GENIE: 100,000 Cases and Beyond.
Pugh TJ, Bell JL, Bruce JP, Doherty GJ, Galvin M, Green MF, Hunter-Zinck H, Kumari P, Lenoue-Newton ML, Li MM, Lindsay J, Mazor T, Ovalle A, Sammut SJ, Schultz N, Yu TV, Sweeney SM, Bernard B; AACR Project GENIE Consortium, Genomics and Analysis Working Group. Pugh TJ, et al. Cancer Discov. 2022 Sep 2;12(9):2044-2057. doi: 10.1158/2159-8290.CD-21-1547. Cancer Discov. 2022. PMID: 35819403 Free PMC article. - AACR releases large cancer genomic data set from project GENIE.
Printz C. Printz C. Cancer. 2017 May 15;123(10):1685. doi: 10.1002/cncr.30755. Cancer. 2017. PMID: 28475245 No abstract available. - American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: From Inception to First Data Release and Beyond-Lessons Learned and Member Institutions' Perspectives.
Micheel CM, Sweeney SM, LeNoue-Newton ML, André F, Bedard PL, Guinney J, Meijer GA, Rollins BJ, Sawyers CL, Schultz N, Shaw KRM, Velculescu VE, Levy MA; AACR Project GENIE Consortium. Micheel CM, et al. JCO Clin Cancer Inform. 2018 Dec;2:1-14. doi: 10.1200/CCI.17.00083. JCO Clin Cancer Inform. 2018. PMID: 30652542 Free PMC article. Review. - Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal.
de Bruijn I, Kundra R, Mastrogiacomo B, Tran TN, Sikina L, Mazor T, Li X, Ochoa A, Zhao G, Lai B, Abeshouse A, Baiceanu D, Ciftci E, Dogrusoz U, Dufilie A, Erkoc Z, Garcia Lara E, Fu Z, Gross B, Haynes C, Heath A, Higgins D, Jagannathan P, Kalletla K, Kumari P, Lindsay J, Lisman A, Leenknegt B, Lukasse P, Madela D, Madupuri R, van Nierop P, Plantalech O, Quach J, Resnick AC, Rodenburg SYA, Satravada BA, Schaeffer F, Sheridan R, Singh J, Sirohi R, Sumer SO, van Hagen S, Wang A, Wilson M, Zhang H, Zhu K, Rusk N, Brown S, Lavery JA, Panageas KS, Rudolph JE, LeNoue-Newton ML, Warner JL, Guo X, Hunter-Zinck H, Yu TV, Pilai S, Nichols C, Gardos SM, Philip J; AACR Project GENIE BPC Core Team, AACR Project GENIE Consortium; Kehl KL, Riely GJ, Schrag D, Lee J, Fiandalo MV, Sweeney SM, Pugh TJ, Sander C, Cerami E, Gao J, Schultz N. de Bruijn I, et al. Cancer Res. 2023 Dec 1;83(23):3861-3867. doi: 10.1158/0008-5472.CAN-23-0816. Cancer Res. 2023. PMID: 37668528 Free PMC article. - How to design a national genomic project-a systematic review of active projects.
Kovanda A, Zimani AN, Peterlin B. Kovanda A, et al. Hum Genomics. 2021 Mar 24;15(1):20. doi: 10.1186/s40246-021-00315-6. Hum Genomics. 2021. PMID: 33761998 Free PMC article. Review.
Cited by
- Ex vivo analysis of DNA repair targeting in extreme rare cutaneous apocrine sweat gland carcinoma.
Mäkelä R, Härmä V, Badra Fajardo N, Wells G, Lygerou Z, Sangfelt O, Kononen J, Rantala JK. Mäkelä R, et al. Oncotarget. 2021 May 25;12(11):1100-1109. doi: 10.18632/oncotarget.27961. eCollection 2021 May 25. Oncotarget. 2021. PMID: 34084283 Free PMC article. - Ganciclovir-induced mutations are present in a diverse spectrum of post-transplant malignancies.
Fang H, Yan HHN, Bilardi RA, Flensburg C, Yang H, Barbour JA, Siu HC, Turski M, Chew E, Xu Z, Lam ST, Sharma R, Xu M, Li J, Ip HW, Cheung CYM, Huen MSY, Sweet-Cordero EA, Majewski IJ, Leung SY, Wong JWH. Fang H, et al. Genome Med. 2022 Oct 31;14(1):124. doi: 10.1186/s13073-022-01131-w. Genome Med. 2022. PMID: 36316687 Free PMC article. - Signal Transducer and Activator of Transcription Proteins at the Nexus of Immunodeficiency, Autoimmunity and Cancer.
Liongue C, Sobah ML, Ward AC. Liongue C, et al. Biomedicines. 2023 Dec 23;12(1):45. doi: 10.3390/biomedicines12010045. Biomedicines. 2023. PMID: 38255152 Free PMC article. Review. - Integrated Molecular Characterization of HER2-Low Breast Cancer Using Next Generation Sequencing (NGS).
Merlin JL, Husson M, Sahki N, Gilson P, Massard V, Harlé A, Leroux A. Merlin JL, et al. Biomedicines. 2023 Nov 28;11(12):3164. doi: 10.3390/biomedicines11123164. Biomedicines. 2023. PMID: 38137385 Free PMC article. - Complex Oncological Decision-Making Utilizing Fast-and-Frugal Trees in a Community Setting-Role of Academic and Hybrid Modeling.
Salgia R, Mambetsariev I, Tan T, Schwer A, Pearlstein DP, Chehabi H, Baroz A, Fricke J, Pharaon R, Romo H, Waddington T, Babikian R, Buck L, Kulkarni P, Cianfrocca M, Djulbegovic B, Pal SK. Salgia R, et al. J Clin Med. 2020 Jun 16;9(6):1884. doi: 10.3390/jcm9061884. J Clin Med. 2020. PMID: 32560187 Free PMC article.
References
- Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31:1803–5. - PubMed
- Tannock IF, Hickman JA. Limits to personalized cancer medicine. N Engl J Med. 2016;375:1289–94. - PubMed
- Prasad V. Perspective: The precision-oncology illusion. Nature. 2016;537:S63. - PubMed
- Joyner MJ, Paneth N, Ioannidis JPA. What happens when under-performing big ideas in research become entrenched? JAMA. 2016;316:1355–6. - PubMed
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- K08 CA188615/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P50 CA083639/CA/NCI NIH HHS/United States
- R01 CA190642/CA/NCI NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- U24 CA209851/CA/NCI NIH HHS/United States
- P30 CA006973/CA/NCI NIH HHS/United States
- U10 CA180950/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 HG008100/HG/NHGRI NIH HHS/United States
- U24 CA210950/CA/NCI NIH HHS/United States
- R01 CA121113/CA/NCI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- U01 CA217842/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 CA182587/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources