Antagonism of type I interferon by flaviviruses - PubMed (original) (raw)
Review
Antagonism of type I interferon by flaviviruses
Lisa Miorin et al. Biochem Biophys Res Commun. 2017.
Abstract
The prompt and tightly controlled induction of type I interferon is a central event of the immune defense against viral infection. Flaviviruses comprise a large family of arthropod-borne positive-stranded RNA viruses, many of which represent a serious threat to global human health due to their high rates of morbidity and mortality. All flaviviruses studied so far have been shown to counteract the host's immune response to establish a productive infection and facilitate viral spread. Here, we review the current knowledge on the main strategies that human pathogenic flaviviruses utilize to escape both type I IFN induction and effector pathways. A better understanding of the specific mechanisms by which flaviviruses activate and evade innate immune responses is critical for the development of better therapeutics and vaccines.
Keywords: Flaviviruses; Innate immunity; Interferon antagonism; Type I interferon; Viral innate immune evasion.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
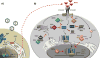
Figure 1. Antagonism of type I IFN production by flaviviruses
Upon recognition of viral RNA, RIG-I and MDA5 undergo a conformational change that triggers their interaction with the adaptor protein MAVS. This in turn induces a signaling cascade that results in IRF3 and NF-kB activation and type I IFN induction (IFN-I) (A). Secreted IFN-I then functions in an autocrine and paracrine fashion leading to the activation of an antiviral state (B). Flaviviruses are able to counteract the induction of type I IFN both in a passive and active fashion. The former functions by hiding flavivirus RNA from PRRs sensing, i.e. by sequestrating dsRNA replication intermediates at the RCs. The later takes place by several mechanisms, unique among different viruses: sfRNAs prevent USP15-mediated deubiquitination of TRIM25, and subsequent RIG-I activation; the NS3 protein of WNV competes with RIG-I for binding to the chaperone 14-3-3ε; the NS4A protein of DENV sequesters MAVS to block downstream signaling events; the structural E protein of WNV inhibits RIP-1 polyubiquitination and blocks the activation of NF-kB signaling. Moreover, DENV infection triggers the release of mtDNA into the cytoplasm. Cytoplasmic mtDNA then binds to cGAS and activates downstream signaling. DENV is able to antagonize the DNA sensing pathway in two ways: the viral protease NS2B3 cleaves STING, and the NS2B protein promotes the degradation of cGAS by an autophagy-lysosome dependent mechanism. Additionally, several nonstructural proteins of both DENV (NS2B3, NS2B, NS2A, NS4A) and WNV (NS4B) have been shown to block the kinases IKKε and TBK-1, that are responsible for IRF3 activation and translocation into the nucleus.
Figure 2. Antagonism of type I IFN signaling by flaviviruses
Mammalian cells sense flavivirus infection mainly via cytoplasmic PRRs such as the RLRs and the DNA sensor cGAS, which in turn activate their adaptor proteins MAVS and STING, respectively. This event initiates downstream signaling cascades ultimately leading to type I IFN (IFN-I) production (A). Once secreted, IFN-I binds to the IFN-I receptor (IFNAR1/IFNAR2) in an autocrine and paracrine manner, and activates the JAK/STAT signaling pathway to trigger an antiviral state (B). Flaviviruses have evolved multiple mechanisms to counteract IFN-I-dependent signaling. JEV NS5 blocks TYK2 phosphorylation. The NS5 proteins of DENV and ZIKV bind to STAT2 and trigger STAT2 proteasomal-degradation. YFV NS5 binds STAT2 upon IFN-I stimulation and TRIM23-dependent ubiquitination. Ubiquitinated YFV NS5 then inhibits ISGF3-driven transcription. SPOV NS5 prevents ISGs transcription in the nucleus trough an uncharacterized mechanism. TBEV and WNV interfere with maturation of the IFNAR1 receptor subunit through the activity of their NS5 proteins. The NS4B proteins of DENV, YFV, and WNV block STAT1 phosphorylation.
Similar articles
- TRIMming down Flavivirus Infections.
Cannac M, Nisole S. Cannac M, et al. Viruses. 2024 Aug 6;16(8):1262. doi: 10.3390/v16081262. Viruses. 2024. PMID: 39205236 Free PMC article. Review. - Tick-Borne Flaviviruses and the Type I Interferon Response.
Lindqvist R, Upadhyay A, Överby AK. Lindqvist R, et al. Viruses. 2018 Jun 21;10(7):340. doi: 10.3390/v10070340. Viruses. 2018. PMID: 29933625 Free PMC article. Review. - Innate Immune DNA Sensing of Flaviviruses.
Zhu T, Fernandez-Sesma A. Zhu T, et al. Viruses. 2020 Sep 3;12(9):979. doi: 10.3390/v12090979. Viruses. 2020. PMID: 32899347 Free PMC article. Review. - Mechanisms of evasion of the type I interferon antiviral response by flaviviruses.
Diamond MS. Diamond MS. J Interferon Cytokine Res. 2009 Sep;29(9):521-30. doi: 10.1089/jir.2009.0069. J Interferon Cytokine Res. 2009. PMID: 19694536 Review. - Innate Immune Antagonism of Mosquito-Borne Flaviviruses in Humans and Mosquitoes.
Elrefaey AME, Hollinghurst P, Reitmayer CM, Alphey L, Maringer K. Elrefaey AME, et al. Viruses. 2021 Oct 20;13(11):2116. doi: 10.3390/v13112116. Viruses. 2021. PMID: 34834923 Free PMC article. Review.
Cited by
- A specific domain within the 3' untranslated region of Usutu virus confers resistance to the exonuclease ISG20.
Zoladek J, El Kazzi P, Caval V, Vivet-Boudou V, Cannac M, Davies EL, Rossi S, Bribes I, Rouilly L, Simonin Y, Jouvenet N, Decroly E, Paillart JC, Wilson SJ, Nisole S. Zoladek J, et al. Nat Commun. 2024 Oct 2;15(1):8528. doi: 10.1038/s41467-024-52870-w. Nat Commun. 2024. PMID: 39358425 Free PMC article. - Innate and adaptive immune responses that control lymph-borne viruses in the draining lymph node.
Melo-Silva CR, Sigal LJ. Melo-Silva CR, et al. Cell Mol Immunol. 2024 Sep;21(9):999-1007. doi: 10.1038/s41423-024-01188-0. Epub 2024 Jun 25. Cell Mol Immunol. 2024. PMID: 38918577 Free PMC article. Review. - ISG15/USP18/STAT2 is a molecular hub regulating IFN I-mediated control of Dengue and Zika virus replication.
Espada CE, da Rocha EL, Ricciardi-Jorge T, Dos Santos AA, Soares ZG, Malaquias G, Patrício DO, Gonzalez Kozlova E, Dos Santos PF, Bordignon J, Sanford TJ, Fajardo T, Sweeney TR, Báfica A, Mansur DS. Espada CE, et al. Front Immunol. 2024 Feb 7;15:1331731. doi: 10.3389/fimmu.2024.1331731. eCollection 2024. Front Immunol. 2024. PMID: 38384473 Free PMC article. - Repurposing of Zika virus live-attenuated vaccine (ZIKV-LAV) strains as oncolytic viruses targeting human glioblastoma multiforme cells.
Victorio CBL, Novera W, Ganasarajah A, Ong J, Thomas M, Wu J, Toh HSY, Sun AX, Ooi EE, Chacko AM. Victorio CBL, et al. J Transl Med. 2024 Feb 2;22(1):126. doi: 10.1186/s12967-024-04930-4. J Transl Med. 2024. PMID: 38308299 Free PMC article. - Mosquito-borne flaviviruses and type I interferon: catch me if you can!
Zoladek J, Nisole S. Zoladek J, et al. Front Microbiol. 2023 Oct 30;14:1257024. doi: 10.3389/fmicb.2023.1257024. eCollection 2023. Front Microbiol. 2023. PMID: 37965539 Free PMC article. Review.
References
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7:787–797. - PubMed
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. - PubMed
- tenOever BR. The Evolution of Antiviral Defense Systems. Cell Host Microbe. 2016;19:142–149. - PubMed
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources