Recent developments in the CCP-EM software suite - PubMed (original) (raw)
Recent developments in the CCP-EM software suite
Tom Burnley et al. Acta Crystallogr D Struct Biol. 2017.
Abstract
As part of its remit to provide computational support to the cryo-EM community, the Collaborative Computational Project for Electron cryo-Microscopy (CCP-EM) has produced a software framework which enables easy access to a range of programs and utilities. The resulting software suite incorporates contributions from different collaborators by encapsulating them in Python task wrappers, which are then made accessible via a user-friendly graphical user interface as well as a command-line interface suitable for scripting. The framework includes tools for project and data management. An overview of the design of the framework is given, together with a survey of the functionality at different levels. The current CCP-EM suite has particular strength in the building and refinement of atomic models into cryo-EM reconstructions, which is described in detail.
Keywords: CCP-EM; Collaborative Computational Project for Electron cryo-Microscopy; cryo-EM.
Figures
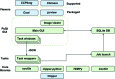
Figure 1
Architecture of the CCP-EM software suite. The task wrappers and core libraries shown in green are written in pure Python, whereas the GUI layer is written in PyQt4. The GUI thread is independent of the job processes; task progress is monitored by a job-launch module and is recorded in an SQLite database. JSON files serve as intermediaries allowing the task to be controlled ‘headless’ without the GUI layer.
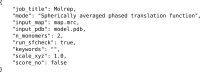
Figure 2
JSON files are used as a convenient, human-readable store of parameters and provide a consistent input for _CCP-EM_-supported applications. In this example, input parameters for a MOLREP job are shown, including the use of a spherically averaged phase translation function and searching for two copies of the search model in the EM volume.
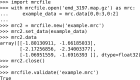
Figure 3
Basic usage of the mrcfile Python library. In this example, a compressed map downloaded from the EMDB is opened and a 2 × 3 slice of data is taken from it. A new MRC file is then created, the data are copied into it and checked, and the file is closed. Finally, the file is validated to confirm that it complies with the MRC2014 standard.
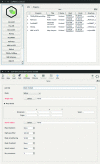
Figure 4
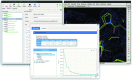
CCP-EM project and task window. Top: CCP-EM project window showing the taskbar which is used to launch applications on the left and the project job history on the right. Bottom: example of the CCP-EM DockEM task. The toolbar at the top gives rapid access to molecular-graphics programs, job files, documentation and job launch. The input parameter setup tab is shown below, with required inputs highlighted in red. Additional launcher and results tabs appear as the job is launched and completed, respectively.
Figure 5
CCP-EM REFMAC task for the refinement and validation of atomic models in high-resolution cryo-EM maps. The single task includes the generation of structure factors from an input reconstruction, as well as the application of multiple blurring and sharpening factors. The left panel shows the CCP-EM pipeline for refinement and validation against experimental half-maps. The centre panel shows the results tab and the right panel shows the input and refined model in Coot.
Figure 6
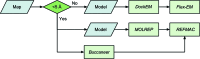
Model-building pipeline in CCP-EM. For maps (or segments thereof) with resolutions of less than ∼5 Å and an appropriate model it is suggested to try DockEM followed by Flex-EM. For higher resolution data MOLREP and REFMAC can be used for refinement if a suitable model is available. If no model is available then Buccaneer can be used to build a model de novo. Note that for medium-resolution data sets a combination of approaches is recommended.
Similar articles
- Overview and applications of map and model validation tools in the CCP-EM software suite.
Joseph AP, Malhotra S, Burnley T, Winn MD. Joseph AP, et al. Faraday Discuss. 2022 Nov 8;240(0):196-209. doi: 10.1039/d2fd00103a. Faraday Discuss. 2022. PMID: 35916020 Free PMC article. Review. - Current approaches for automated model building into cryo-EM maps using Buccaneer with CCP-EM.
Hoh SW, Burnley T, Cowtan K. Hoh SW, et al. Acta Crystallogr D Struct Biol. 2020 Jun 1;76(Pt 6):531-541. doi: 10.1107/S2059798320005513. Epub 2020 May 29. Acta Crystallogr D Struct Biol. 2020. PMID: 32496215 Free PMC article. - Collaborative computational project for electron cryo-microscopy.
Wood C, Burnley T, Patwardhan A, Scheres S, Topf M, Roseman A, Winn M. Wood C, et al. Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):123-6. doi: 10.1107/S1399004714018070. Epub 2015 Jan 1. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25615866 Free PMC article. - Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM.
Nicholls RA, Tykac M, Kovalevskiy O, Murshudov GN. Nicholls RA, et al. Acta Crystallogr D Struct Biol. 2018 Jun 1;74(Pt 6):492-505. doi: 10.1107/S2059798318007313. Epub 2018 May 30. Acta Crystallogr D Struct Biol. 2018. PMID: 29872001 Free PMC article. Review. - cryoem-cloud-tools: A software platform to deploy and manage cryo-EM jobs in the cloud.
Cianfrocco MA, Lahiri I, DiMaio F, Leschziner AE. Cianfrocco MA, et al. J Struct Biol. 2018 Sep;203(3):230-235. doi: 10.1016/j.jsb.2018.05.014. Epub 2018 Jun 1. J Struct Biol. 2018. PMID: 29864529 Free PMC article.
Cited by
- Structural basis for DNA recognition by a viral genome-packaging machine.
Chechik M, Greive SJ, Antson AA, Jenkins HT. Chechik M, et al. Proc Natl Acad Sci U S A. 2024 Aug 13;121(33):e2406138121. doi: 10.1073/pnas.2406138121. Epub 2024 Aug 8. Proc Natl Acad Sci U S A. 2024. PMID: 39116131 Free PMC article. - Structural basis for the multitasking nature of the potato virus Y coat protein.
Kežar A, Kavčič L, Polák M, Nováček J, Gutiérrez-Aguirre I, Žnidarič MT, Coll A, Stare K, Gruden K, Ravnikar M, Pahovnik D, Žagar E, Merzel F, Anderluh G, Podobnik M. Kežar A, et al. Sci Adv. 2019 Jul 17;5(7):eaaw3808. doi: 10.1126/sciadv.aaw3808. eCollection 2019 Jul. Sci Adv. 2019. PMID: 31328164 Free PMC article. - A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, Ding S, Benlarbi M, Gong SY, Tauzin A, Gasser R, Chatterjee D, Vézina D, Goyette G, Richard J, Zhou F, Stamatatos L, McGuire AT, Charest H, Roger M, Pozharski E, Kumar P, Mothes W, Uchil PD, Pazgier M, Finzi A. Beaudoin-Bussières G, et al. Cell Rep. 2022 Feb 15;38(7):110368. doi: 10.1016/j.celrep.2022.110368. Epub 2022 Jan 25. Cell Rep. 2022. PMID: 35123652 Free PMC article. - Protein dynamics developments for the large scale and cryoEM: case study of ProDy 2.0.
Krieger JM, Sorzano COS, Carazo JM, Bahar I. Krieger JM, et al. Acta Crystallogr D Struct Biol. 2022 Apr 1;78(Pt 4):399-409. doi: 10.1107/S2059798322001966. Epub 2022 Mar 16. Acta Crystallogr D Struct Biol. 2022. PMID: 35362464 Free PMC article. Review. - Overview and applications of map and model validation tools in the CCP-EM software suite.
Joseph AP, Malhotra S, Burnley T, Winn MD. Joseph AP, et al. Faraday Discuss. 2022 Nov 8;240(0):196-209. doi: 10.1039/d2fd00103a. Faraday Discuss. 2022. PMID: 35916020 Free PMC article. Review.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
- Andreani, J. & Söding, J. (2015). Bioinformatics, 31, 1729–1737. - PubMed
- Biyani, N., Righetto, R. D., McLeod, R., Caujolle-Bert, D., Castano-Diez, D., Goldie, K. N. & Stahlberg, H. (2017). J. Struct. Biol. 198, 124–133. - PubMed
- Briggs, P. (2007). CCP4 Newsl. Protein Crystallogr. 46, contribution 5.
MeSH terms
Grants and funding
- BB/P000975/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/J000825/1/MRC_/Medical Research Council/United Kingdom
- MR/N009614/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials