Mechanistic elucidation of the mycofactocin-biosynthetic radical S-adenosylmethionine protein, MftC - PubMed (original) (raw)
Mechanistic elucidation of the mycofactocin-biosynthetic radical _S_-adenosylmethionine protein, MftC
Bulat Khaliullin et al. J Biol Chem. 2017.
Abstract
Ribosomally synthesized and posttranslationally modified peptide (RiPP) pathways produce a diverse array of natural products. A subset of these pathways depends on radical _S_-adenosylmethionine proteins to modify the RiPP-produced peptide. Mycofactocin biosynthesis is one example of an _S_-adenosylmethionine protein-dependent RiPP pathway. Recently, it has been shown that MftC catalyzes the oxidative decarboxylation of the C-terminal tyrosine (Tyr-30) on the mycofactocin precursor peptide MftA; however, this product has not been verified by techniques other than MS. Herein, we provide a more detailed study of MftC catalysis and report a revised mechanism for MftC chemistry. We show that MftC catalyzes the formation of two isomeric products. Using a combination of MS, isotope labeling, and 1H and 13C NMR techniques, we established that the major product, MftA*, is a tyramine-valine-cross-linked peptide formed by MftC through two _S_-adenosylmethionine-dependent turnovers. In addition, we show that the hydroxyl group on MftA Tyr-30 is required for MftC catalysis. Furthermore, we show that a substitution in the penultimate MftA Val-29 position causes the accumulation of an MftA** minor product. The 1H NMR spectrum indicates that this minor product contains an αβ-unsaturated bond that likely arises from an aborted intermediate of MftA* synthesis. The finding that MftA* is the major product formed during MftC catalysis could have implications for the further elucidation of mycofactocin biosynthesis.
Keywords: MftC; S-adenosylmethionine (SAM); enzyme mechanism; iron-sulfur protein; mycofactocin; peptide biosynthesis; radical.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Figure 1.
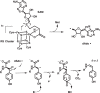
Previously proposed reaction mechanism for MftC.
Figure 2.
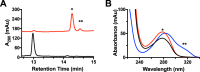
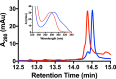
MftC catalyzed the formation of two MftA products annotated as MftA* and MftA**. A, HPLC analysis of the reactions shows the conversion of MftA M1W (black) to MftA* (14.3 min) and MftA** (14.5 min). Analyses were carried out on a C4 HPLC column. B, the absorbance spectra of MftA M1W (black) and MftA* (red) are similar, whereas the absorbance spectra for MftA** (blue) indicates the appearance of a new spectral feature from 300–325 nm and a red-shift in peak maximum. mAu, milliabsorbance units. *, MftA*; **, MftA**.
Figure 3.
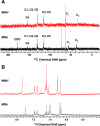
13C (A) and 1H (B) NMR analyses of MftA* did not indicate that an αβ-unsaturated bond is present. MftA is represented by black spectra, and MftA* is represented by red spectra.
Figure 4.
Analysis of the mass spectrum of Val-29 (D8)-labeled MftA M1W-purified peptide (A) and Val-29 (D8)-labeled MftA M1W* (B) indicated that Val-29 forms a new intramolecular bond. Mass spectra of Val-29 (3-D)-labeled MftA M1W (C) and Val-29 (3-D) labeled MftA M1W* (D) indicates that the new intramolecular bond is formed on the Cβ of Val-29. All collected spectral data are represented in black, and the predicted spectra for the deuterium-labeled MftA MH3+ ions are shown in red. The predicted spectra for unlabeled MftA MH3+ is shown in blue for panels A and B.
Figure 5.
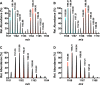
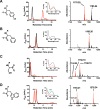
The MftA M1W/V29A mutant led to the accumulation of MftA M1W/V29A** product. HPLC analysis of the reactions shows the conversion of MftA M1W/V29A (black) to MftA* (13.7 min) and MftA** (14.1 min). Analyses were carried out on a C4 HPLC column. B, the absorbance spectra of MftA M1W/V29A (black), MftA M1W/V29A* (red), and MftA M1W/V29A** (blue) were similar to what was found for reactions with MftA M1W. C, 1H NMR analysis of MftA M1W/V29A** demonstrated the formation of an αβ-unsaturated indicated by appearance of doublets at ∼6.3 and ∼7.2 ppm (indicated by **). mAu, milliabsorbance units.
Figure 6.
Shown are HRMS analyses of dAdo control (A), simulated incorporation of one deuterium in dAdo (B), dAdo isolated from a reaction with Tyr-30 (ring-2,6-D2, 2-D) MftA (Cα-labeled) (C), dAdo isolated from a reaction with Tyr-30 (3,3-D2) MftA (Cβ-labeled) (D), and dAdo isolated from a reaction with Val-29 (3-D) MftA (Cβ-labeled) (E).
Figure 7.
MftC catalyzed the conversion of MftA** to MftA*. HPLC analysis of MftA** starting material (blue) and of a reaction with MftC (red) indicates a change in retention time and absorbance spectrum (inset). Analyses were carried out on a C4 HPLC column. mAu, milliabsorbance units.
Figure 8.
HPLC, UV-visible, and HRMS analysis of MftA M1W (A), MftA M1W/Y30F (B), MftA M1W/Y30S (C), and MftA M1W/Y30W (D). Each variant has a corresponding HPLC chromatogram (middle column), a UV-visible spectrum (middle column, inset), and an ion chromatogram (right column). HPLC analyses were carried out on a C4 HPLC column. For all HPLC and HRMS chromatograms, black is the unreacted MftA variant, and red represents the reaction with the MftA variant. For the UV-visible spectra, black is the unreacted MftA variant, red is the reacted MftA variant, and cyan is dAdo adduct to the Y30W variant. mAu, milliabsorbance units.
Figure 9.
Revised mechanism for MftC catalysis.
Similar articles
- Mycofactocin biosynthesis: modification of the peptide MftA by the radical S-adenosylmethionine protein MftC.
Khaliullin B, Aggarwal P, Bubas M, Eaton GR, Eaton SS, Latham JA. Khaliullin B, et al. FEBS Lett. 2016 Aug;590(16):2538-48. doi: 10.1002/1873-3468.12249. Epub 2016 Jun 29. FEBS Lett. 2016. PMID: 27312813 - Spectroscopic and Electrochemical Characterization of the Mycofactocin Biosynthetic Protein, MftC, Provides Insight into Its Redox Flipping Mechanism.
Ayikpoe R, Ngendahimana T, Langton M, Bonitatibus S, Walker LM, Eaton SS, Eaton GR, Pandelia ME, Elliott SJ, Latham JA. Ayikpoe R, et al. Biochemistry. 2019 Feb 19;58(7):940-950. doi: 10.1021/acs.biochem.8b01082. Epub 2019 Jan 25. Biochemistry. 2019. PMID: 30628436 Free PMC article. - Mycofactocin Biosynthesis Proceeds through 3-Amino-5-[( p-hydroxyphenyl)methyl]-4,4-dimethyl-2-pyrrolidinone (AHDP); Direct Observation of MftE Specificity toward MftA.
Ayikpoe R, Salazar J, Majestic B, Latham JA. Ayikpoe R, et al. Biochemistry. 2018 Sep 18;57(37):5379-5383. doi: 10.1021/acs.biochem.8b00816. Epub 2018 Sep 6. Biochemistry. 2018. PMID: 30183269 Free PMC article. - Occurrence, function, and biosynthesis of mycofactocin.
Ayikpoe R, Govindarajan V, Latham JA. Ayikpoe R, et al. Appl Microbiol Biotechnol. 2019 Apr;103(7):2903-2912. doi: 10.1007/s00253-019-09684-4. Epub 2019 Feb 18. Appl Microbiol Biotechnol. 2019. PMID: 30778644 Free PMC article. Review. - Radical S-Adenosylmethionine Enzymes Involved in RiPP Biosynthesis.
Mahanta N, Hudson GA, Mitchell DA. Mahanta N, et al. Biochemistry. 2017 Oct 10;56(40):5229-5244. doi: 10.1021/acs.biochem.7b00771. Epub 2017 Sep 22. Biochemistry. 2017. PMID: 28895719 Free PMC article. Review.
Cited by
- Sequence-function space of radical SAM cyclophane synthases reveal conserved active site residues that influence substrate specificity.
Phan CS, Morinaka BI. Phan CS, et al. RSC Chem Biol. 2024 Oct 23;5(12):1195-200. doi: 10.1039/d4cb00227j. Online ahead of print. RSC Chem Biol. 2024. PMID: 39464308 Free PMC article. - Decarboxylation in Natural Products Biosynthesis.
Nguyen NA, Forstater JH, McIntosh JA. Nguyen NA, et al. JACS Au. 2024 Jul 25;4(8):2715-2745. doi: 10.1021/jacsau.4c00425. eCollection 2024 Aug 26. JACS Au. 2024. PMID: 39211618 Free PMC article. Review. - Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Lien Y, Lachowicz JC, Mendauletova A, Zizola C, Ngendahimana T, Kostenko A, Eaton SS, Latham JA, Grove TL. Lien Y, et al. ACS Chem Biol. 2024 Feb 16;19(2):370-379. doi: 10.1021/acschembio.3c00583. Epub 2024 Jan 31. ACS Chem Biol. 2024. PMID: 38295270 - Nocardioides jiangxiensis sp. nov., a novel actinobacterium isolated from lakeside soil, exhibiting the biosynthesis potential of mycofactocin.
Wan Y, Sun T, Huang G, Tu B, Huang M, Chen X, Liu B, He J. Wan Y, et al. Antonie Van Leeuwenhoek. 2024 Jan 8;117(1):18. doi: 10.1007/s10482-023-01910-4. Antonie Van Leeuwenhoek. 2024. PMID: 38190009 - A mycofactocin-associated dehydrogenase is essential for ethylene glycol metabolism by Rhodococcus jostii RHA1.
Shimizu T, Suzuki K, Inui M. Shimizu T, et al. Appl Microbiol Biotechnol. 2024 Dec;108(1):58. doi: 10.1007/s00253-023-12966-7. Epub 2024 Jan 4. Appl Microbiol Biotechnol. 2024. PMID: 38175243
References
- Haft D. H., Pierce P. G., Mayclin S. J., Sullivan A., Gardberg A. S., Abendroth J., Begley D. W., Phan I. Q., Staker B. L., Myler P. J., Marathias V. M., Lorimer D. D., and Edwards T. E. (2017) Mycofactocin-associated mycobacterial dehydrogenases with non-exchangeable NAD cofactors. Sci. Rep. 7, 41074. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases