Fast and accurate HLA typing from short-read next-generation sequence data with xHLA - PubMed (original) (raw)
. 2017 Jul 25;114(30):8059-8064.
doi: 10.1073/pnas.1707945114. Epub 2017 Jul 3.
Zhen Xuan Yeo 2, Marie Wong 2, Jason Piper 2, Tao Long 3, Ewen F Kirkness 3, William H Biggs 3, Ken Bloom 3, Stephen Spellman 4, Cynthia Vierra-Green 4, Colleen Brady 4, Richard H Scheuermann 5 6, Amalio Telenti 3, Sally Howard 3, Suzanne Brewerton 2, Yaron Turpaz 2 3, J Craig Venter 7 5
Affiliations
- PMID: 28674023
- PMCID: PMC5544337
- DOI: 10.1073/pnas.1707945114
Fast and accurate HLA typing from short-read next-generation sequence data with xHLA
Chao Xie et al. Proc Natl Acad Sci U S A. 2017.
Abstract
The HLA gene complex on human chromosome 6 is one of the most polymorphic regions in the human genome and contributes in large part to the diversity of the immune system. Accurate typing of HLA genes with short-read sequencing data has historically been difficult due to the sequence similarity between the polymorphic alleles. Here, we introduce an algorithm, xHLA, that iteratively refines the mapping results at the amino acid level to achieve 99-100% four-digit typing accuracy for both class I and II HLA genes, taking only [Formula: see text]3 min to process a 30× whole-genome BAM file on a desktop computer.
Keywords: MHC; autoimmune diseases; transplantation.
Conflict of interest statement
Conflict of interest statement: C.X., Z.X.Y., M.W., J.P., T.L., E.F.K., W.H.B., K.B., A.T., S.H., S.B., Y.T., and J.C.V. are employees of Human Longevity Inc. There is a patent pending on the technique presented in this work.
Figures
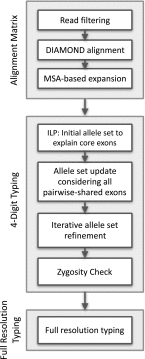
Fig. 1.
Overview of the xHLA algorithm.
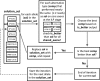
Fig. 2.
Iterative allele set update considering all pairwise-shared exons.
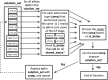
Fig. 3.
Second-round iterative allele set refinement.
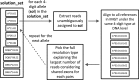
Fig. 4.
Full-resolution HLA typing as a downstream step after four-digit typing.
Similar articles
- Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms.
Liu P, Yao M, Gong Y, Song Y, Chen Y, Ye Y, Liu X, Li F, Dong H, Meng R, Chen H, Zheng A. Liu P, et al. Front Immunol. 2021 Mar 31;12:652258. doi: 10.3389/fimmu.2021.652258. eCollection 2021. Front Immunol. 2021. PMID: 33868290 Free PMC article. - HLA-HD: An accurate HLA typing algorithm for next-generation sequencing data.
Kawaguchi S, Higasa K, Shimizu M, Yamada R, Matsuda F. Kawaguchi S, et al. Hum Mutat. 2017 Jul;38(7):788-797. doi: 10.1002/humu.23230. Epub 2017 May 12. Hum Mutat. 2017. PMID: 28419628 - Benchmarking freely available HLA typing algorithms across varying genes, coverages and typing resolutions.
Thuesen NH, Klausen MS, Gopalakrishnan S, Trolle T, Renaud G. Thuesen NH, et al. Front Immunol. 2022 Nov 8;13:987655. doi: 10.3389/fimmu.2022.987655. eCollection 2022. Front Immunol. 2022. PMID: 36426357 Free PMC article. - Histocompatibility testing.
Chee RE. Chee RE. Southeast Asian J Trop Med Public Health. 2002;33 Suppl 2:112-4. Southeast Asian J Trop Med Public Health. 2002. PMID: 12755280 Review.
Cited by
- Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms.
Liu P, Yao M, Gong Y, Song Y, Chen Y, Ye Y, Liu X, Li F, Dong H, Meng R, Chen H, Zheng A. Liu P, et al. Front Immunol. 2021 Mar 31;12:652258. doi: 10.3389/fimmu.2021.652258. eCollection 2021. Front Immunol. 2021. PMID: 33868290 Free PMC article. - Strength of immune selection in tumors varies with sex and age.
Castro A, Pyke RM, Zhang X, Thompson WK, Day CP, Alexandrov LB, Zanetti M, Carter H. Castro A, et al. Nat Commun. 2020 Aug 17;11(1):4128. doi: 10.1038/s41467-020-17981-0. Nat Commun. 2020. PMID: 32807809 Free PMC article. - Strategies for neoantigen screening and immunogenicity validation in cancer immunotherapy (Review).
Feng H, Jin Y, Wu B. Feng H, et al. Int J Oncol. 2025 Jun;66(6):43. doi: 10.3892/ijo.2025.5749. Epub 2025 May 9. Int J Oncol. 2025. PMID: 40342048 Free PMC article. Review. - Clonal replacement of tumor-specific T cells following PD-1 blockade.
Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, Curtis C, Bucktrout SL, Davis MM, Chang ALS, Chang HY. Yost KE, et al. Nat Med. 2019 Aug;25(8):1251-1259. doi: 10.1038/s41591-019-0522-3. Epub 2019 Jul 29. Nat Med. 2019. PMID: 31359002 Free PMC article. - Bioinformatics Strategies, Challenges, and Opportunities for Next Generation Sequencing-Based HLA Genotyping.
Klasberg S, Surendranath V, Lange V, Schöfl G. Klasberg S, et al. Transfus Med Hemother. 2019 Oct;46(5):312-325. doi: 10.1159/000502487. Epub 2019 Sep 6. Transfus Med Hemother. 2019. PMID: 31832057 Free PMC article. Review.
References
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: Expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. - PubMed
- Robinson J, Soormally AR, Hayhurst JD, Marsh SGE. The IPD-IMGT/HLA Database—new developments in reporting HLA variation. Hum Immunol. 2016;77:233–237. - PubMed
- Prugnolle F, et al. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15:1022–1027. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous