Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis - PubMed (original) (raw)
doi: 10.1038/s41467-017-00050-4.
Marghoob Mohiyuddin 1, Robert Sebra 2, Hagen Tilgner 3, Pegah T Afshar 4, Kin Fai Au 5, Narges Bani Asadi 1, Mark B Gerstein 6, Wing Hung Wong 7, Michael P Snyder 3, Eric Schadt 2, Hugo Y K Lam 8
Affiliations
- PMID: 28680106
- PMCID: PMC5498581
- DOI: 10.1038/s41467-017-00050-4
Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis
Sayed Mohammad Ebrahim Sahraeian et al. Nat Commun. 2017.
Abstract
RNA-sequencing (RNA-seq) is an essential technique for transcriptome studies, hundreds of analysis tools have been developed since it was debuted. Although recent efforts have attempted to assess the latest available tools, they have not evaluated the analysis workflows comprehensively to unleash the power within RNA-seq. Here we conduct an extensive study analysing a broad spectrum of RNA-seq workflows. Surpassing the expression analysis scope, our work also includes assessment of RNA variant-calling, RNA editing and RNA fusion detection techniques. Specifically, we examine both short- and long-read RNA-seq technologies, 39 analysis tools resulting in ~120 combinations, and ~490 analyses involving 15 samples with a variety of germline, cancer and stem cell data sets. We report the performance and propose a comprehensive RNA-seq analysis protocol, named RNACocktail, along with a computational pipeline achieving high accuracy. Validation on different samples reveals that our proposed protocol could help researchers extract more biologically relevant predictions by broad analysis of the transcriptome.RNA-seq is widely used for transcriptome analysis. Here, the authors analyse a wide spectrum of RNA-seq workflows and present a comprehensive analysis protocol named RNACocktail as well as a computational pipeline leveraging the widely used tools for accurate RNA-seq analysis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
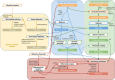
Fig. 1
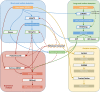
The RNACocktail analysis protocol. RNACocktail is a comprehensive protocol of RNA-seq data analysis. The figure summarizes the widely used approaches for the key steps over the broad spectrum of RNA-seq analysis and also succinctly captures the possible workflows one can use to analyse RNA-seq data
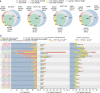
Fig. 2
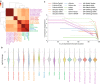
Performance of different alignment schemes. a Overlap between the detected splice junctions by different schemes and their validation rate on reliable junctions in dbEST database. A reliable EST junction set consists of junctions supported by at least two ESTs. The sizes of the circles reflect the number of junctions called by each scheme. For each tool, the number of junctions called and the validation rates (in parentheses) are shown. Validation rates for each subset of junctions are also shown on the Venn diagram. b Read mapping analysis: distribution of mapping status of sequenced fragments (left) (for NA12878, MCF7, and SEQC samples, mapping status for paired-end reads are shown, while for hESC, the distribution reflects percentage of uniquely mapped (blue), multi-mapped (orange), and unmapped (red) single-end reads), distribution of number of soft-clipped bases in mapped fragments (middle), distribution of the number of mismatches in mapped fragments (right)
Fig. 3
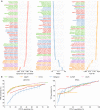
Performance of different transcriptome reconstruction schemes. a Distribution of number of exons per transcripts for different transcriptome reconstruction algorithms. Labels reflect the assembler, the long-read aligner (for IDP), and the short-read aligner used, respectively, with “-” separation. b Sensitivity and precision of different transcriptome reconstruction approaches at gene and transcript levels. The GENCODE reference transcriptome annotation is used as the truth set. The evaluations on a more recent update of MCF7 sample using the Iso-Seq pipeline resulted in a similar performance with only slight improvement. The union approaches that combined predictions from short reads and long reads (shown with a “+” in the label) slightly improved the performance of short-read isoform prediction schemes
Fig. 4
Performance of different de novo transcriptome assembly techniques. a Distribution of transcript length. b N10-N50 values. (Nx is the contig length for which at least x% of the assembled transcript nucleotides were found in contigs that were at least of Nx length.) c ExN50 value at different expression percentiles. For ExN50, the N50 statistic was computed at percentile x, for only the top most highly expressed transcripts that represent x% of the total normalized expression data. Expression values were measured using eXpress
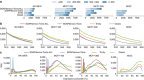
Fig. 5
Performance of transcript abundance estimators. a Clustering of different schemes based on the Spearman rank correlation of their log expressions on NA12878. b Distribution of log2-fold change of expressions between MCF7-100 and MCF7-300 samples. For each method, dashed line represents the mean of the distribution and the dotted lines represents the quartiles. c Percentage of expression disagreement between MCF7-100 and MCF7-300 samples when low-expressed transcripts are discarded with different thresholds
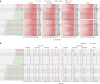
Fig. 6
Performance of differential gene expressions analysis tools on SEQC-A vs. SEQC-B samples. a Spearman rank correlation, root-mean-score-deviation (RMSD), and AUC-30 scores for qPCR measured genes. Spearman rank correlation and RMSD scores are measured between the log2-fold change of the qRT-PCR and RNA-seq tools. AUC-30 score represents the area under the ROC curve up to the false positive rate of 30%. b ROC analysis of qRT-PCR measured genes (left) and ERCC (right) genes. For each differential analysis tool the plot reflects average performance when different alignment-based and alignment-free tools are used for abundance estimation and error bar shows the maximum and minimum variations. Results for each tool combination are shown in Supplementary Figs. 30 and 35
Fig. 7
Performance of different variant calling (a–c) RNA editing (d, e) and RNA fusion (f) detection approaches. a Accuracy of detecting NA12878 high-confidence calls in NIST gold standard. The analysis is restricted to the expressed exons identified by Cufflinks or StringTie. b Overlap between variants predicted by GATK and SAMtools. c Distribution of the predicted mismatch types by GATK that are missed in NIST HC calls (in StringTie’s expressed exons) in different genomic regions. d Distribution of RNA editing events detected in different genomic regions for NA12878. For multiple-samples scheme, final editing sites include the rare variants in NA12878 that are supported by at least 3 out of 12 short-read samples in our analysis. For pooled-samples scheme, final editing sites include the rare variants in NA12878 that are supported by at least 20 reads in the pooled alignment. e Measurement of RNA editing detection accuracy when some portion of SNPs are hidden from GIREMI. FDR represent proportion of predicted edits that are among high-confidence genomic variants in NA12878. f Performance of different RNA fusion detection schemes on MCF-7 sample
Fig. 8
The current RNACocktail computational pipeline. The pipeline is composed of high-accuracy tools in each step for general-purpose RNA-seq analysis
Similar articles
- Automated Isoform Diversity Detector (AIDD): a pipeline for investigating transcriptome diversity of RNA-seq data.
Plonski NM, Johnson E, Frederick M, Mercer H, Fraizer G, Meindl R, Casadesus G, Piontkivska H. Plonski NM, et al. BMC Bioinformatics. 2020 Dec 30;21(Suppl 18):578. doi: 10.1186/s12859-020-03888-6. BMC Bioinformatics. 2020. PMID: 33375933 Free PMC article. - Empirical assessment of analysis workflows for differential expression analysis of human samples using RNA-Seq.
Williams CR, Baccarella A, Parrish JZ, Kim CC. Williams CR, et al. BMC Bioinformatics. 2017 Jan 17;18(1):38. doi: 10.1186/s12859-016-1457-z. BMC Bioinformatics. 2017. PMID: 28095772 Free PMC article. - Read-Split-Run: an improved bioinformatics pipeline for identification of genome-wide non-canonical spliced regions using RNA-Seq data.
Bai Y, Kinne J, Donham B, Jiang F, Ding L, Hassler JR, Kaufman RJ. Bai Y, et al. BMC Genomics. 2016 Aug 22;17 Suppl 7(Suppl 7):503. doi: 10.1186/s12864-016-2896-7. BMC Genomics. 2016. PMID: 27556805 Free PMC article. - Characterizing and annotating the genome using RNA-seq data.
Chen G, Shi T, Shi L. Chen G, et al. Sci China Life Sci. 2017 Feb;60(2):116-125. doi: 10.1007/s11427-015-0349-4. Epub 2016 Jun 13. Sci China Life Sci. 2017. PMID: 27294835 Review. - Overview of available methods for diverse RNA-Seq data analyses.
Chen G, Wang C, Shi T. Chen G, et al. Sci China Life Sci. 2011 Dec;54(12):1121-8. doi: 10.1007/s11427-011-4255-x. Epub 2012 Jan 7. Sci China Life Sci. 2011. PMID: 22227904 Review.
Cited by
- The need for masked genomes in gymnosperms.
Zhu P, He T, Zheng Y, Chen L. Zhu P, et al. Front Plant Sci. 2023 Dec 7;14:1309744. doi: 10.3389/fpls.2023.1309744. eCollection 2023. Front Plant Sci. 2023. PMID: 38146270 Free PMC article. No abstract available. - Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome.
Fu S, Tang X, Xu Y, Song X, Qian X, Hu Y, Zhang M. Fu S, et al. Life (Basel). 2022 Nov 23;12(12):1961. doi: 10.3390/life12121961. Life (Basel). 2022. PMID: 36556326 Free PMC article. - Analysis of public RNA-sequencing data reveals biological consequences of genetic heterogeneity in cell line populations.
Fasterius E, Al-Khalili Szigyarto C. Fasterius E, et al. Sci Rep. 2018 Jul 25;8(1):11226. doi: 10.1038/s41598-018-29506-3. Sci Rep. 2018. PMID: 30046134 Free PMC article. - Comprehensive Analysis of Large-Scale Transcriptomes from Multiple Cancer Types.
Nong B, Guo M, Wang W, Songyang Z, Xiong Y. Nong B, et al. Genes (Basel). 2021 Nov 24;12(12):1865. doi: 10.3390/genes12121865. Genes (Basel). 2021. PMID: 34946814 Free PMC article. - Transcriptomic response to three osmotic stresses in gills of hybrid tilapia (Oreochromis mossambicus female × O. urolepis hornorum male).
Su H, Ma D, Zhu H, Liu Z, Gao F. Su H, et al. BMC Genomics. 2020 Jan 31;21(1):110. doi: 10.1186/s12864-020-6512-5. BMC Genomics. 2020. PMID: 32005144 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases