Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity - PubMed (original) (raw)
Review
Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity
Vinod K Gupta et al. Front Microbiol. 2017.
Abstract
One of the fundamental issues in the microbiome research is characterization of the healthy human microbiota. Recent studies have elucidated substantial divergences in the microbiome structure between healthy individuals from different race and ethnicity. This review provides a comprehensive account of such geography, ethnicity or life-style-specific variations in healthy microbiome at five major body habitats-Gut, Oral-cavity, Respiratory Tract, Skin, and Urogenital Tract (UGT). The review focuses on the general trend in the human microbiome evolution-a gradual transition in the gross compositional structure along with a continual decrease in diversity of the microbiome, especially of the gut microbiome, as the human populations passed through three stages of subsistence like foraging, rural farming and industrialized urban western life. In general, gut microbiome of the hunter-gatherer populations is highly abundant with Prevotella, Proteobacteria, Spirochaetes, Clostridiales, Ruminobacter etc., while those of the urban communities are often enriched in Bacteroides, Bifidobacterium, and Firmicutes. The oral and skin microbiome are the next most diverse among different populations, while respiratory tract and UGT microbiome show lesser variations. Higher microbiome diversity is observed for oral-cavity in hunter-gatherer group with higher prevalence of Haemophilus than agricultural group. In case of skin microbiome, rural and urban Chinese populations show variation in abundance of Trabulsiella and Propionibacterium. On the basis of published data, we have characterized the core microbiota-the set of genera commonly found in all populations, irrespective of their geographic locations, ethnicity or mode of subsistence. We have also identified the major factors responsible for geography-based alterations in microbiota; though it is not yet clear which factor plays a dominant role in shaping the microbiome-nature or nurture, host genetics or his environment. Some of the geographical/racial variations in microbiome structure have been attributed to differences in host genetics and innate/adaptive immunity, while in many other cases, cultural/behavioral features like diet, hygiene, parasitic load, environmental exposure etc. overshadow genetics. The ethnicity or population-specific variations in human microbiome composition, as reviewed in this report, question the universality of the microbiome-based therapeutic strategies and recommend for geographically tailored community-scale approaches to microbiome engineering.
Keywords: body habitats; disease susceptibility; host genetics; hunter-gatherers; lifestyle; non-western people; rural community; urban life.
Figures
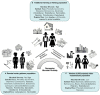
Figure 1
Enriched taxa at various niches of the human body in diverse populations around the world. Box color: body niche; Color in Map: percentage urbanization of countries (
); Up arrow: Dominant abundance of Phylum/Genus compared to respective population; Down arrow: Low abundances of Phylum/Genus/family compared to respective population; * and # comparisons between specific countries; Number in respective boxes: Citations.
Figure 2
Gradual transition of the gut microbiota composition with changes in the host subsistence strategies.
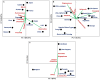
Figure 3
Principal Component Analysis based on relative abundances of core microbiota among different countries/populations derived from a specific body niche (population wise data shown in Tables S1–S3): (A) Gut (B) Oral cavity (C) Vagina.
Similar articles
- Variations in the Post-weaning Human Gut Metagenome Profile As Result of Bifidobacterium Acquisition in the Western Microbiome.
Soverini M, Rampelli S, Turroni S, Schnorr SL, Quercia S, Castagnetti A, Biagi E, Brigidi P, Candela M. Soverini M, et al. Front Microbiol. 2016 Jul 12;7:1058. doi: 10.3389/fmicb.2016.01058. eCollection 2016. Front Microbiol. 2016. PMID: 27462302 Free PMC article. - Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet.
Lassalle F, Spagnoletti M, Fumagalli M, Shaw L, Dyble M, Walker C, Thomas MG, Bamberg Migliano A, Balloux F. Lassalle F, et al. Mol Ecol. 2018 Jan;27(1):182-195. doi: 10.1111/mec.14435. Epub 2017 Dec 14. Mol Ecol. 2018. PMID: 29165844 - Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence.
Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, Blekhman R, Ségurel L. Morton ER, et al. PLoS Genet. 2015 Nov 30;11(11):e1005658. doi: 10.1371/journal.pgen.1005658. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26619199 Free PMC article. - Human behavior, not race or geography, is the strongest predictor of microbial succession in the gut bacteriome of infants.
Quin C, Gibson DL. Quin C, et al. Gut Microbes. 2020 Sep 2;11(5):1143-1171. doi: 10.1080/19490976.2020.1736973. Epub 2020 Apr 5. Gut Microbes. 2020. PMID: 32249675 Free PMC article. Review. - Population-specific differences in the human microbiome: Factors defining the diversity.
Govender P, Ghai M. Govender P, et al. Gene. 2025 Jan 15;933:148923. doi: 10.1016/j.gene.2024.148923. Epub 2024 Sep 6. Gene. 2025. PMID: 39244168 Review.
Cited by
- Impact of Daily Consumption of Whole-Grain Quinoa-Enriched Bread on Gut Microbiome in Males.
Li L, Houghton D, Lietz G, Watson A, Stewart CJ, Bal W, Seal CJ. Li L, et al. Nutrients. 2022 Nov 18;14(22):4888. doi: 10.3390/nu14224888. Nutrients. 2022. PMID: 36432574 Free PMC article. Clinical Trial. - Codiversification of gut microbiota with humans.
Suzuki TA, Fitzstevens JL, Schmidt VT, Enav H, Huus KE, Mbong Ngwese M, Grießhammer A, Pfleiderer A, Adegbite BR, Zinsou JF, Esen M, Velavan TP, Adegnika AA, Song LH, Spector TD, Muehlbauer AL, Marchi N, Kang H, Maier L, Blekhman R, Ségurel L, Ko G, Youngblut ND, Kremsner P, Ley RE. Suzuki TA, et al. Science. 2022 Sep 16;377(6612):1328-1332. doi: 10.1126/science.abm7759. Epub 2022 Sep 15. Science. 2022. PMID: 36108023 Free PMC article. - Learning machine approach reveals microbial signatures of diet and sex in dog.
Scarsella E, Stefanon B, Cintio M, Licastro D, Sgorlon S, Dal Monego S, Sandri M. Scarsella E, et al. PLoS One. 2020 Aug 17;15(8):e0237874. doi: 10.1371/journal.pone.0237874. eCollection 2020. PLoS One. 2020. PMID: 32804973 Free PMC article. - Eating Patterns and Dietary Interventions in ADHD: A Narrative Review.
Pinto S, Correia-de-Sá T, Sampaio-Maia B, Vasconcelos C, Moreira P, Ferreira-Gomes J. Pinto S, et al. Nutrients. 2022 Oct 16;14(20):4332. doi: 10.3390/nu14204332. Nutrients. 2022. PMID: 36297016 Free PMC article. Review. - Associations of healthy food choices with gut microbiota profiles.
Koponen KK, Salosensaari A, Ruuskanen MO, Havulinna AS, Männistö S, Jousilahti P, Palmu J, Salido R, Sanders K, Brennan C, Humphrey GC, Sanders JG, Meric G, Cheng S, Inouye M, Jain M, Niiranen TJ, Valsta LM, Knight R, Salomaa VV. Koponen KK, et al. Am J Clin Nutr. 2021 Aug 2;114(2):605-616. doi: 10.1093/ajcn/nqab077. Am J Clin Nutr. 2021. PMID: 34020448 Free PMC article.
References
- Albert A. Y., Chaban B., Wagner E. C., Schellenberg J. J., Links M. G., van Schalkwyk J., et al. . (2015). A study of the vaginal microbiome in healthy canadian women utilizing cpn60-based molecular profiling reveals distinct gardnerella subgroup community state types. PLoS ONE 10:e0135620. 10.1371/journal.pone.0135620 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources