Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation - PubMed (original) (raw)
doi: 10.1038/s41598-017-06016-2.
Chieko Saito 2 3, Miki Nakazawa 4, Shozo Fujioka 5, Tomohiro Uemura 3, Minami Matsui 6, Masaaki Sakuta 7, Kazuo Shinozaki 1, Hiroyuki Osada 5, Akihiko Nakano 2 3 8, Tadao Asami 1 9 10 11, Takeshi Nakano 12 13
Affiliations
- PMID: 28720789
- PMCID: PMC5515986
- DOI: 10.1038/s41598-017-06016-2
Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation
Ayumi Yamagami et al. Sci Rep. 2017.
Abstract
Brassinosteroids (BRs), plant steroid hormones, play important roles in plant cell elongation and differentiation. To investigate the mechanisms of BR signaling, we previously used the BR biosynthesis inhibitor Brz as a chemical biology tool and identified the Brz-insensitive-long hypocotyl4 mutant (bil4). Although the BIL4 gene encodes a seven-transmembrane-domain protein that is evolutionarily conserved in plants and animals, the molecular function of BIL4 in BR signaling has not been elucidated. Here, we demonstrate that BIL4 is expressed in early elongating cells and regulates cell elongation in Arabidopsis. BIL4 also activates BR signaling and interacts with the BR receptor brassinosteroid insensitive 1 (BRI1) in endosomes. BIL4 deficiency increases the localization of BRI1 in the vacuoles. Our results demonstrate that BIL4 regulates cell elongation and BR signaling via the regulation of BRI1 localization.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Figure 1
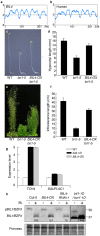
BIL4 is a positive regulator of BR signaling. (a,b) Hydrophobicity profiles of BIL4 (a) and the human homolog Golgi anti-apoptotic protein (hGAAP) (b). (c,d) Phenotype (c) and hypocotyl length (d) of wild-type (WT), bri1-5 and BIL4-OX bri1-5 double mutant seedlings grown in the dark for 7 days. The results are presented as the mean ± s.d. (n > 30 seedlings). (e,f) Phenotype (e) and inflorescence length (f) of WT, bri1-5 and BIL4-OX bri1-5 plants grown in soil for 6 weeks. The results are presented as the mean ± s.d. (n > 20 plants). (g) qRT-PCR analyses of TCH4 and SAUR-AC1 expression levels in wild-type, bil4-1D, and BIL4-OX plants grown in the dark for 8 days. The results are presented as the mean ± s.d. (h) BIL1/BZR1 phosphorylation status in wild-type, BIL4-OX, BIL4-RNAi-1 and bil1-1D/bzr1-1D plants (pBIL1/BZR1, phospho-BIL1/BZR1). Plants that were grown on medium containing 3 µM Brz in the dark for 7 days were submerged for 3 hr in medium containing 100 nM BL. Western blot analyses were performed using the anti-BIL1/BZR1 antibody (upper panel). The protein levels were detected using Ponceau S (lower panel). Numbers indicate the relative BIL1/BZR1 signal levels normalized to the Ponceau S-stained protein band.
Figure 2
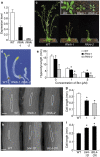
BIL4 is important for cell elongation. (a) qRT-PCR analysis of BIL4 expression in wild-type (WT), BIL4-RNAi-1 (RNAi-1) and BIL4-RNAi-2 (RNAi-2) seedlings grown in the dark for 3 days. The results are presented as the mean ± s.d. (b,c) Three-week-old (b) and 5-week-old (c) wild-type and BIL4-RNAi plants. (d,e) Phenotype (d) and hypocotyl length (e) of wild-type and BIL4-RNAi seedlings grown on medium containing 3 µM Brz in the dark for 6 days. The results are presented as the mean ± s.d., n > 30 seedlings. Asterisks indicate a significant difference from the wild-type plant (_P <_ 0.01 by Student’s _t_-test). (**f** to **i**) SEM images (**f,h**) and length (**g,i**) of the hypocotyl cells. The typical cell phenotype is marked by a white frame. Scale bar, 100 µm. The results are presented as the mean ± s.e.m. Asterisks indicate a significant difference relative to wild-type plants (_P <_ 0.01 by Student’s _t_-test). _RNAi_ seedlings were germinated on 1 µM Brz (n > 17 cells; f,g), and overexpressing seedlings were germinated on 3 µM Brz (n > 35 cells; h and i) in the dark for 7 days.
Figure 3
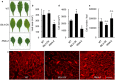
BIL4 positively affects the size of leaf epidermal cells. (a) Rosette leaves of wild-type, BIL4-OX and BIL4-RNAi-2 (RNAi-2) plants 6 weeks after sowing. (b) Leaf area of wild-type, BIL4-OX and RNAi-2 plants. The results are presented as the mean ± s.d., n = 5 leaves. Asterisks indicate a significant difference from the wild-type plants (P < 0.01 by Student’s _t_-test). (c) Confocal images of the epidermis of 6-week-old wild-type, BIL4-OX and RNAi-2 rosette leaves stained with PI. Scale bar, 50 μm. (d) Cell size in the epidermis of wild-type, BIL4-OX and RNAi-2 rosette leaves. The results are presented as the mean ± s.d., n = 9 areas. Asterisks indicate a significant difference from the wild-type plant (P < 0.01 by Student’s _t_-test). (e) Epidermal cell number of wild-type, BIL4-OX and RNAi-2 rosette leaves. The results are presented as the mean ± s.d., n = 5 leaves; n.s. indicates no significant difference from the wild-type plants (by Student’s _t_-test).
Figure 4
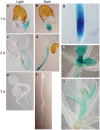
BIL4 is specifically expressed during the activation of cell elongation. (a–f) BIL4 promoter (BIL4pro)::GUS expression pattern 1 (a), 2 (c) and 3 (e) days after germination in the light. BIL4pro::GUS expression pattern 1 (b), 2 (d) and 3 (f) days after germination in the dark. (g–i) BIL4pro::GUS is expressed in the roots (g), very small rosette leaves (h) and the short bolting stem (i).
Figure 5
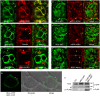
BIL4 is localized to punctate structures and the vacuolar membrane and interacts with BRI1. (a) _BIL4_pro::BIL4–GFP partially co-localizes with the vacuolar membrane marker mRFP–VAM3 (unmerged puncta are marked by white arrows). (b) _BIL4_pro::BIL4–GFP partially colocalizes with FM4-64. Seedlings were treated with FM4-64 for 5 min and then incubated in water for 40 min. (c) Seedlings were pretreated with FM4-64 for 5 min, incubated in water for 20 min and then treated with 50 μM BFA for 20 min. (d–f) _BIL4_pro::BIL4–GFP partially co-localizes with the TGN/EE marker mRFP–SYP41 (d), the LE/MVB marker mRFP–ARA7 (e) and _BRI1_pro::BRI1–Venus in the endosome (f). Merged structures in b–f are marked by yellow arrows. Scale bar in (a–f): 5 µm. (g) BiFC assay of the interactions between BIL4 and BRI1 in cultured Arabidopsis cells. Scale bars, 10 μm. The white arrowheads indicate characteristic interactions. (h) Co-immunoprecipitation of BRI1 and BIL4. Wild-type (Col-0) and transgenic plants coexpressing _BRI1_pro::BRI1–GFP and 35 S::FLAG–BIL4 were grown for 7 days. FLAG–BIL4 was immunoprecipitated by anti-FLAG antibody, and the immunoblots were probed with anti-GFP or anti-FLAG antibody.
Figure 6
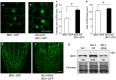
BRI1 subcellular localization is affected in the BIL4-OX and BIL4-RNAi plants. (a–d) Three-day-old seedlings were treated with BFA (50 μM, 0.5 hr). BRI1–GFP-labeled BFA bodies in the wild-type (a) and BIL4-OX plants (b). Scale bar, 10 µm. Signal intensities of BRI1–GFP-labeled BFA bodies in the wild-type and _BIL4_-OX plants (c). Sizes of BRI1–GFP-labeled BFA bodies in the wild-type and BIL4-OX plants (d). (c,d) n = 3 roots with at least 30 BFA bodies. *P < 0.01, Student’s _t_-test. Mean ± s.e. (e,f) BRI1–GFP localization in the root tip of wild-type (e) and BIL4-RNAi mutant (f) 2 days after germination in the dark. Scale bar, 10 µm. (g) Plants were grown in the dark for 7 days on medium containing 3 µM Brz. Western blot analyses were performed using the anti-BRI1 antibody (upper panel). The protein levels were detected using Ponceau S (lower panel). Numbers indicate the relative BRI1 signal levels normalized to the Ponceau S-stained protein band.
Similar articles
- Chemical genetics reveal the novel transmembrane protein BIL4, which mediates plant cell elongation in brassinosteroid signaling.
Yamagami A, Nakazawa M, Matsui M, Tujimoto M, Sakuta M, Asami T, Nakano T. Yamagami A, et al. Biosci Biotechnol Biochem. 2009 Feb;73(2):415-21. doi: 10.1271/bbb.80752. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202280 - Brassinosteroids regulate vacuolar morphology in root meristem cells of Arabidopsis thaliana.
Yamagami A, Chieko S, Sakuta M, Shinozaki K, Osada H, Nakano A, Asami T, Nakano T. Yamagami A, et al. Plant Signal Behav. 2018 Apr 3;13(4):e1417722. doi: 10.1080/15592324.2017.1417722. Epub 2018 Apr 16. Plant Signal Behav. 2018. PMID: 29252095 Free PMC article. - Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1.
Jiang J, Zhang C, Wang X. Jiang J, et al. J Integr Plant Biol. 2013 Dec;55(12):1198-211. doi: 10.1111/jipb.12081. Epub 2013 Sep 9. J Integr Plant Biol. 2013. PMID: 23718739 Review. - Immunophilin-like FKBP42/TWISTED DWARF1 Interacts with the Receptor Kinase BRI1 to Regulate Brassinosteroid Signaling in Arabidopsis.
Chaiwanon J, Garcia VJ, Cartwright H, Sun Y, Wang ZY. Chaiwanon J, et al. Mol Plant. 2016 Apr 4;9(4):593-600. doi: 10.1016/j.molp.2016.01.008. Epub 2016 Jan 22. Mol Plant. 2016. PMID: 26808213 Free PMC article. - Brassinosteroid signaling in plant development and adaptation to stress.
Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. Planas-Riverola A, et al. Development. 2019 Mar 14;146(5):dev151894. doi: 10.1242/dev.151894. Development. 2019. PMID: 30872266 Free PMC article. Review.
Cited by
- Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway.
Mao J, Li J. Mao J, et al. Int J Mol Sci. 2020 Jun 18;21(12):4340. doi: 10.3390/ijms21124340. Int J Mol Sci. 2020. PMID: 32570783 Free PMC article. Review. - Whether Gametophytes are Reduced or Unreduced in Angiosperms Might Be Determined Metabolically.
Mateo de Arias M, Gao L, Sherwood DA, Dwivedi KK, Price BJ, Jamison M, Kowallis BM, Carman JG. Mateo de Arias M, et al. Genes (Basel). 2020 Dec 2;11(12):1449. doi: 10.3390/genes11121449. Genes (Basel). 2020. PMID: 33276690 Free PMC article. - Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum).
Lu R, Zhang J, Liu D, Wei YL, Wang Y, Li XB. Lu R, et al. BMC Plant Biol. 2018 Nov 27;18(1):304. doi: 10.1186/s12870-018-1523-y. BMC Plant Biol. 2018. PMID: 30482177 Free PMC article. - Arabidopsis zinc finger homeodomain transcription factor BRASSINOSTEROID-RELATED HOMEOBOX 2 acts as a positive regulator of brassinosteroid response.
Hasegawa R, Fujita K, Tanaka Y, Takasaki H, Ikeda M, Yamagami A, Mitsuda N, Nakano T, Ohme-Takagi M. Hasegawa R, et al. Plant Biotechnol (Tokyo). 2022 Jun 25;39(2):185-189. doi: 10.5511/plantbiotechnology.22.0115a. Plant Biotechnol (Tokyo). 2022. PMID: 35937534 Free PMC article. - Arabidopsis homeobox-leucine zipper transcription factor BRASSINOSTEROID-RELATED HOMEOBOX 3 regulates leaf greenness by suppressing BR signaling.
Hasegawa R, Arakawa T, Fujita K, Tanaka Y, Ookawa Z, Sakamoto S, Takasaki H, Ikeda M, Yamagami A, Mitsuda N, Nakano T, Ohme-Takagi M. Hasegawa R, et al. Plant Biotechnol (Tokyo). 2022 Jun 25;39(2):209-214. doi: 10.5511/plantbiotechnology.22.0128a. Plant Biotechnol (Tokyo). 2022. PMID: 35937537 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases