CD300f:IL-5 cross-talk inhibits adipose tissue eosinophil homing and subsequent IL-4 production - PubMed (original) (raw)
CD300f:IL-5 cross-talk inhibits adipose tissue eosinophil homing and subsequent IL-4 production
Perri Rozenberg et al. Sci Rep. 2017.
Abstract
Eosinophils and their associated cytokines IL-4 and IL-5 are emerging as central orchestrators of the immune-metabolic axis. Herein, we demonstrate that cross-talk between the Ig-superfamily receptor CD300f and IL-5 is a key checkpoint that modifies the ability of eosinophils to regulate metabolic outcomes. Generation of Il5 Tg /Cd300f -/- mice revealed marked and distinct increases in eosinophil levels and their production of IL-4 in the white and brown adipose tissues. Consequently, Il5 Tg /Cd300f -/- mice had increased alternatively activated macrophage accumulation in the adipose tissue. Cd300f -/- mice displayed age-related accumulation of eosinophils and macrophages in the adipose tissue and decreased adipose tissue weight, which was associated with decreased diet-induced weight gain and insulin resistance. Notably, Il5 Tg /CD300f -/- were protected from diet-induced weight gain and glucose intolerance. These findings highlight the cross-talk between IL-5 receptor and CD300f as a novel pathway regulating adipose tissue eosinophils and offer new entry points for therapeutic intervention for obesity and its complications.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Figure 1
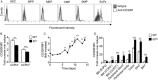
CD300f is expressed by eosinophil progenitors and regulated by IL-5. The expression of CD300f was assessed in bone marrow hematopoietic stem cells (HSCs, CD45+/Lin−/Sca-1−/c-kit+/CD135−), multipotent progenitors (MPPs, CD45+/Lin−/Sca-1−/c-kit+/CD135−), myeloid-erythrocyte progenitors (MEPs, CD45+/Lin−/Sca-1+/c-kit+/CD34−/CD32/16+), common myeloid progenitors (CMPs, CD45+/Lin−/Sca-1+/c-kit+/CD34+/CD32/16−), granulocyte-macrophage progenitor (GMPs, CD45+/Lin−/Sca-1+/c-kit+/CD34+/CD16+) and eosinophil progenitors (EoPs, CD45+/Lin−/Sca-1+/c-kit+/CD34int/IL-5Rα+) (A). The expression of CD300f was assessed in immature (Siglec-F+/CCR3−) and mature (Siglec-F+/CCR3+) bone marrow eosinophils in wild type (WT) and Il5 −/− mice (B). Bone marrow-derived eosinophils were generated ex-vivo and the expression of CD300f on Siglec-F+ cells was determined at the indicated time points throughout the eosinophil cell culture (C). Finally, the expression of CD300f was assessed in eosinophils (Eos) (D), from the indicated organs and in adipose tissue macrophages (Mac), monocytes (Mono) and neutrophils (Neut) (E) that were obtained from WT and hypereosinophilic Il5 transgenic (Il5 Tg) mice (D,E). In (E), the red line indicates fold-increase of 1. Data for (A) are representative of n = 5 mice, for (B), n = 6 mice, for (C), representative of 3–4 independent repeats; for (D), n = 3–4 mice; *p < 0.05, **p < 0.01, ***p < 0.001 as analyzed by Student’s t-test (B and D) and two-way ANOVA followed by Tukey post-hoc test (C).
Figure 2
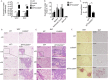
IL-5-driven adipose tissue eosinophil accumulation is negatively regulated by CD300f. The number of total eosinophils was assessed in the white adipose tissue (WAT) and brown adipose tissue (BAT) of six to eight-week old wild type (WT), Cd300f −/−, Il5 Tg and Il5 Tg /Cd300f −/− mice (A). In addition, eosinophil levels were enumerated in the bone marrow (BM), spleen, peritoneal cavity, jejunum (B) and peripheral blood (C) of Il5 Tg and Il5 Tg /Cd300f −/− mice (B). The WAT (D, F) and BAT (E,F) of wild type (WT), Cd300f −/−, Il5 Tg and Il5 Tg /Cd300f −/− mice was obtained and subjected to H&E staining (D,E) as well as anti-eosinophil major basic protein (MBP) stain (F). Data for (A–C) are from n = 5 mice. In (D), representative photomicrographs are shown; **p < 0.01, ***p < 0.001 as analyzed by Student’s t-test (A and C).
Figure 3
CD300f negatively regulates eosinophil-derived IL-4 production and governs IL-5-induced ERK and pAKT phosphorylation. The mRNA (A) expression of IL-4 was assessed in the white and brown adipose tissue (WAT and BAT, respectively) by quantitative PCR and normalized to the house keeping gene hypoxanthine-guanine phosphoribosyltransferase (Hprt). Protein expression of IL-4 (B) was determined by ELISA. In (C), primary eosinophils were purified from the WAT of Il5 Tg and Il5 Tg /Cd300f −/− mice and either left untreated (NT) or activated with phorbol 12- myristate 13-acetate (PMA). Thereafter, secretion of IL-4 was determined in the culture supernatants (C). The expression of CD300f (D) and IL-5 receptor α (IL-5Rα) (G) in I.29 B cells is shown following viral infection with empty vector or CD300f-containing vector (D). Following retroviral infection with empty vector of CD300f-containg vector, I.29 cells were stimulated with IL-5 for the indicated time points (E,F) and the phosphorylation of ERK (p42/44) (E,F) and AKT (H) was assessed by phosphoflow (E,F) and western blot, respectively (H). Data in (A,B) are from n = 5 mice, in (C) from n = 3 independent experiments, in (D,H) from n = 4 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001 as analyzed by Student’s t-test (A–C) and two-way Anova followed by Tukey post-hoc test (F).
Figure 4
CD300f-IL-5 receptor cross talk regulates alternatively activated macrophage formation in adipose tissue. The expression of hallmark alternatively activated macrophage markers such as Arg1, Chi3l, Relm-α and Ccl24 were determined in the adipose tissue of wild type (WT), Cd300f −/−, Il5 Tg and Il5 Tg /Cd300f −/− mice by quantitative PCR and normalized to the house keeping gene hypoxanthine-guanine phosphoribosyltransferase (Hprt) (A,B and D) and ELISA (C). Furthermore, the expression of Arg1, Chi3l, and Ccl24 was determined in the primary adipose tissue macrophages that were sorted from Il5 Tg and Il5 Tg /Cd300f −/− mice by quantitative PCR (E–G). In situ proliferation of adipose tissue eosinophils, macrophages and monocytes was assessed in Il5 Tg and Il5 Tg /Cd300f −/− mice using flow cytometric analysis of EdU+ cells (H). Thereafter, total WAT and BAT macrophage levels in Il5 Tg and Il5 Tg /Cd300f −/− mice were assessed (I). Data in (A–D) are from n = 5–6 mice; in (E–G) n = 3; in (H,I) from n = 6; *p < 0.05, **p < 0.01, ***p < 0.001 as analyzed by Student’s t-test (A–I).
Figure 5
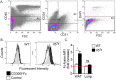
Viable adipose tissue endothelial cells express a CD300f ligand. White adipose tissue (A–D) and lungs (D) from wild type (WT) and Il5 Tg mice were obtained, enzymatically digested and stained with viability and endothelial cells markers (A). Thereafter, the binding of CD300f-IgG1 Fc fusion protein to viable adipose tissue (B,C) and lung (D) endothelial cells was determined by flow cytometry. Data are representative of n = 4 independent experiments; *p < 0.05, ns- non-significant as analyzed by Student’s t-test (C).
Figure 6
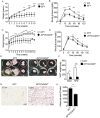
Cd300f −/− mice display age-related accumulation of eosinophils and macrophages and decreased adipose tissue weight. White adipose tissue (WAT) of young (2–4 months) and aged (5–8 months) wild type (WT) and Cd300f −/− mice were obtained and total eosinophils (A) and macrophages (B) per mg tissue were determined. Moreover, the weight of WATs from WT and Cd300f −/− mice was determined (C). In (D) a representative photograph of WAT size from WT and Cd300f −/− mice is shown. WT and Cd300f −/− mice were fed with high fat diet (HFD) for up to 18 weeks and weighed at the indicated time points (E). Following 18 weeks of HFD, mice were tested for glucose tolerance (F). Data in (A–C) are from n = 3, 2–3 mice per time point; in (D) data are representative of on one out of eight; in (E–F) data are from n = 3 using 10 mice per group; *p < 0.05, **p < 0.01, ***p < 0.001 as analyzed by Student’s t-test (A–C) and two-way ANOVA followed by Tukey post-hoc test (E,F).
Figure 7
Il5 Tg /CD300f −/− mice are protected from diet-induced weight gain and glucose intolerance. Wild type (WT), Il5 Tg and Il5 Tg /CD300f −/− mice were fed with high fat diet (HFD) for up to 12 weeks and their weight was monitored at the indicated time points (A,C). After ten (B) and twelve (D) weeks of HFD, the mice underwent glucose tolerance test (B,D). Il5 Tg and Il5 Tg /CD300f −/− mice were fed with NCD or HFD for twelve weeks. Thereafter, the white adipose tissue (WAT) was obtained, photographed (E) and weighed (F). In addition, adipocyte size was assessed in H&E stained slides (G–H). Data in (A–D) are representative of n = 12; in (E and G) photographs are representative of 6/12 mice; In (F and H), n = 14; *p < 0.05, **p < 0.01, ***p < 0.001 as analyzed by two-way ANOVA followed by Tukey post-hoc test (A–D) and Student’s t-test (F–H).
Similar articles
- CD300f associates with IL-4 receptor α and amplifies IL-4-induced immune cell responses.
Moshkovits I, Karo-Atar D, Itan M, Reichman H, Rozenberg P, Morgenstern-Ben-Baruch N, Shik D, Ejarque-Ortiz A, Hershko AY, Tian L, Coligan JE, Sayós J, Munitz A. Moshkovits I, et al. Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8708-13. doi: 10.1073/pnas.1507625112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124135 Free PMC article. - CCR2 deficiency leads to increased eosinophils, alternative macrophage activation, and type 2 cytokine expression in adipose tissue.
Bolus WR, Gutierrez DA, Kennedy AJ, Anderson-Baucum EK, Hasty AH. Bolus WR, et al. J Leukoc Biol. 2015 Oct;98(4):467-77. doi: 10.1189/jlb.3HI0115-018R. Epub 2015 May 1. J Leukoc Biol. 2015. PMID: 25934927 Free PMC article. - Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis.
Wu D, Molofsky AB, Liang HE, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, Chawla A, Locksley RM. Wu D, et al. Science. 2011 Apr 8;332(6026):243-7. doi: 10.1126/science.1201475. Epub 2011 Mar 24. Science. 2011. PMID: 21436399 Free PMC article. - [Growth and differentiation of eosinophils--special reference to IL-5 and its receptor].
Akutsu I, Takatsu K. Akutsu I, et al. Nihon Rinsho. 1993 Mar;51(3):557-64. Nihon Rinsho. 1993. PMID: 8492427 Review. Japanese. - CD300 family receptors regulate eosinophil survival, chemotaxis, and effector functions.
Rozenberg P, Reichman H, Moshkovits I, Munitz A. Rozenberg P, et al. J Leukoc Biol. 2018 Jul;104(1):21-29. doi: 10.1002/JLB.2MR1117-433R. Epub 2017 Dec 21. J Leukoc Biol. 2018. PMID: 29345367 Review.
Cited by
- Eosinophils in obesity and obesity-associated disorders.
Hu Y, Chakarov S. Hu Y, et al. Discov Immunol. 2023 Nov 14;2(1):kyad022. doi: 10.1093/discim/kyad022. eCollection 2023. Discov Immunol. 2023. PMID: 38567054 Free PMC article. Review. - Energy Metabolism, Metabolite, and Inflammatory Profiles in Human Ex Vivo Adipose Tissue Are Influenced by Obesity Status, Metabolic Dysfunction, and Treatment Regimes in Patients with Oesophageal Adenocarcinoma.
O'Connell F, Mylod E, Donlon NE, Heeran AB, Butler C, Bhardwaj A, Ramjit S, Durand M, Lambe G, Tansey P, Welartne I, Sheahan KP, Yin X, Donohoe CL, Ravi N, Dunne MR, Brennan L, Reynolds JV, Roche HM, O'Sullivan J. O'Connell F, et al. Cancers (Basel). 2023 Mar 9;15(6):1681. doi: 10.3390/cancers15061681. Cancers (Basel). 2023. PMID: 36980567 Free PMC article. - Rebamipide treatment ameliorates obesity phenotype by regulation of immune cells and adipocytes.
Jhun J, Moon J, Kim SY, Cho KH, Na HS, Choi J, Jung YJ, Song KY, Min JK, Cho ML. Jhun J, et al. PLoS One. 2022 Dec 27;17(12):e0277692. doi: 10.1371/journal.pone.0277692. eCollection 2022. PLoS One. 2022. PMID: 36574392 Free PMC article. - Differential regulation of Type 1 and Type 2 mouse eosinophil activation by apoptotic cells.
Dolitzky A, Hazut I, Avlas S, Grisaru-Tal S, Itan M, Zaffran I, Levi-Schaffer F, Gerlic M, Munitz A. Dolitzky A, et al. Front Immunol. 2022 Oct 31;13:1041660. doi: 10.3389/fimmu.2022.1041660. eCollection 2022. Front Immunol. 2022. PMID: 36389786 Free PMC article. - Immune Cells in Thermogenic Adipose Depots: The Essential but Complex Relationship.
Agueda-Oyarzabal M, Emanuelli B. Agueda-Oyarzabal M, et al. Front Endocrinol (Lausanne). 2022 Mar 14;13:839360. doi: 10.3389/fendo.2022.839360. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35360060 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous