The N-terminal dimerization is required for TDP-43 splicing activity - PubMed (original) (raw)
The N-terminal dimerization is required for TDP-43 splicing activity
Lei-Lei Jiang et al. Sci Rep. 2017.
Abstract
TDP-43 is a nuclear factor that functions in promoting pre-mRNA splicing. Deletion of the N-terminal domain (NTD) and nuclear localization signal (NLS) (i.e., TDP-35) results in mislocalization to cytoplasm and formation of inclusions. However, how the NTD functions in TDP-43 activity and proteinopathy remains largely unknown. Here, we studied the structure and function of the NTD in inclusion formation and pre-mRNA splicing of TDP-43 by using biochemical and biophysical approaches. We found that TDP-43 NTD forms a homodimer in solution in a concentration-dependent manner, and formation of intermolecular disulfide results in further tetramerization. Based on the NMR structure of TDP-43 NTD, the dimerization interface centered on Leu71 and Val72 around the β7-strand was defined by mutagenesis and size-exclusion chromatography. Cell experiments revealed that the N-terminal dimerization plays roles in protecting TDP-43 against formation of cytoplasmic inclusions and enhancing pre-mRNA splicing activity of TDP-43 in nucleus. This study may provide mechanistic insights into the physiological function of TDP-43 and its related proteinopathies.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Figure 1
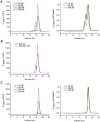
Characterization of the oligomeric states of the N-terminal fragments of TDP-43 by SEC. (A) Schematic representation of the domain architecture of TDP-43 and its N-terminal fragments. NTD, N-terminal domain; NLS, nuclear localization signal; RRM, RNA recognition motif; GRR, glycine-rich region; AC, amyloidogenic core. (B) SEC analysis of the N-terminal fragments of TDP-43. The proteins were diluted with Buffer A (25 mM Tris-HCl, pH 8.0, and 150 mM NaCl) to a concentration of 50 µM and loaded onto a Superdex-200 Increase 10/30 GL column. The chromatographic profiles are shown for TDP(1–261) (black), TDP(1–89) (red), and TDP(101–261) (blue). A280nm, Absorbance at 280 nm; mAU, micro absorbance unit. (C) Standard profile for calculating the apparent molecular weight (MW). Ve/V0, relative elution volume. The standard markers are: BSA (67.0 kDa), GST (52.6 kDa), ovobumin (44.3 kDa), thioredoxin (13.9 kDa), and cytochrome C (12.4 kDa). The concentrations of the standards are around 10 to 100 μM. (D) As in (B), SEC profiles of the N-terminal fragments of TDP-43 at 10-µM concentration.
Figure 2
SEC profiles for TDP(1–77) and its Cys mutant. (A) SEC profiles for TDP(1–77) at different concentrations. (B) SEC profiles for TDP(1–77) with (pink) or without 5 mM DTT (black). The protein concentration was to 500 µM. (C) SEC profiles for TDP(1–77)-C39/C50S at different concentrations. The SEC experiments were carried out using a Superdex-200 Increase 10/30 GL column previously equilibrated with Buffer A. The right panels show the normalized chromatograms.
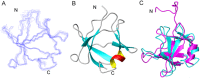
Figure 3
Solution structure of TDP(1–77) (C39/C50S) elucidated by NMR. (A) Superposition of the backbone traces of the 10 lowest-energy structures. (B) Ribbon diagram of a representative structure of TDP(1–77) (C39/C50S), showing seven β-strands and one short α-helix. The structure was elucidated with the GB1-fused Cys mutant TDP(1–77)-GB1-C39/C50S. (C) Comparison of the overall structures of TDP(1–77) from TDP(1–77)-GB1-C39/C50S (blue) and His6-TDP(1–77) (pink; PDB: 2N4P). The RMSD is 2.96 Å. N, N-terminus; C, C-terminus. All the structures are displayed using MOLMOL.
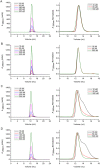
Figure 4
Identification of the dimer interfaces of TDP(1–77) by mutagenesis and SEC. (A) SEC profiles for TDP(1–77)-GB1-C39/C50S. (B) L27/I60/V57A mutant. (C) L71/Y73A mutant. (D) V72/V74/Y43A mutant. The right panels show the normalized chromatograms. The mutants were generated based on TDP(1–77)-GB1-C39/C50S. The proteins were diluted to the indicated concentrations with Buffer A and loaded onto a Superdex-75 10/30 GL column.
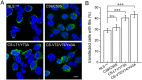
Figure 5
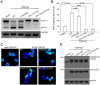
Immunofluorescence microscopic imaging of cytoplasmic inclusions of TDP-43-NLSmut and its dimer-interface mutants. (A) Inclusion formation of FLAG-TDP-43-NLSmut and its Cys mutant (C39/C50S) and dimer-interface mutants (CS-L71/Y73A and CS-V72/V74/Y43A) in HEK 293 T cells. TDP-43-NLSmut and its mutants were stained with mouse anti-FLAG (green), and the nuclei were stained with Hoechst (blue). The images are shown with a magnification of 3 times (3 X). Scale bar = 10 µm. The mutants were generated based on FLAG-TDP-43-NLSmut. (B) Quantification of the cells with cytoplasmic inclusions in HEK 293 T cells overexpressing FLAG-TDP-43-NLSmut or its mutants. Cells were counted and the data were statistically analyzed by one-way ANOVA. Results are presented as Mean ± SEM (n = 25–30). ***p < 0.001; n.s., no significance.
Figure 6
Effects of dimer-interface mutations on the RNA splicing activity of TDP-43. (A) Effects of TDP-43, its Cys mutant (C39/C50S) and dimer-interface mutants (CS-L71/Y73A and CS-V72/V74/Y43A) on CFTR exon 9 splicing. The GT13T5 reporter was co-transfected with the FLAG-tagged plasmids into HEK 293 T cells, respectively. Exon 9 + stands for unspliced cDNA fragment, while exon 9− denotes spliced cDNA. (B) Quantification of the spliced RT-PCR products of TDP-43 and its mutants based on band intensity. Data were analyzed by one-way ANOVA and represented as Mean ± SD (n = 3). ***p < 0.001. (C) Immunofluorescence microscopic images showing nuclear localization of FLAG-TDP-43 and its mutants in HEK 293 T cells. TDP-43 and its mutants were stained with mouse anti-TDP-43 or anti-FLAG (green), and the nuclei were stained with Hoechst (blue). The images are shown with a magnification of 3 times (3 X). Scale bar = 10 µm. (D) Western blot showing the expression levels of the protein species for CFTR splicing assay.
Figure 7
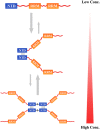
Schematic representation for the oligomerization of TDP-43. The model proposes that TDP-43 NTD mainly forms a dimer via residues Leu71 and Val72 around the β7-strand, and then transforms into a tetramer via disulfide formation at high protein concentrations. The NTD dimerization or tetramerization may protect TDP-43 against aggregation and is required for splicing activity of TDP-43.
Similar articles
- Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.
Mompeán M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, Laurents DV. Mompeán M, et al. J Biol Chem. 2017 Jul 14;292(28):11992-12006. doi: 10.1074/jbc.M117.775965. Epub 2017 May 31. J Biol Chem. 2017. PMID: 28566288 Free PMC article. - A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing.
Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, Shewmaker F, Naik MT, Mittag T, Ayala YM, Fawzi NL. Wang A, et al. EMBO J. 2018 Mar 1;37(5):e97452. doi: 10.15252/embj.201797452. Epub 2018 Feb 9. EMBO J. 2018. PMID: 29438978 Free PMC article. - Insight into the Folding and Dimerization Mechanisms of the N-Terminal Domain from Human TDP-43.
Vivoli-Vega M, Guri P, Chiti F, Bemporad F. Vivoli-Vega M, et al. Int J Mol Sci. 2020 Aug 29;21(17):6259. doi: 10.3390/ijms21176259. Int J Mol Sci. 2020. PMID: 32872449 Free PMC article. - TDP-43: a DNA and RNA binding protein with roles in neurodegenerative diseases.
Warraich ST, Yang S, Nicholson GA, Blair IP. Warraich ST, et al. Int J Biochem Cell Biol. 2010 Oct;42(10):1606-9. doi: 10.1016/j.biocel.2010.06.016. Epub 2010 Jun 25. Int J Biochem Cell Biol. 2010. PMID: 20601083 Review. - [The molecular mechanisms of intracellular TDP-43 aggregates].
Nonaka T, Arai T, Hasegawa M. Nonaka T, et al. Brain Nerve. 2009 Nov;61(11):1292-300. Brain Nerve. 2009. PMID: 19938686 Review. Japanese.
Cited by
- Conformational Enigma of TDP-43 Misfolding in Neurodegenerative Disorders.
Pillai M, Jha SK. Pillai M, et al. ACS Omega. 2024 Sep 20;9(39):40286-40297. doi: 10.1021/acsomega.4c04119. eCollection 2024 Oct 1. ACS Omega. 2024. PMID: 39372031 Free PMC article. Review. - Elucidation of the structural stability and dynamics of heterogeneous intermediate ensembles in unfolding pathway of the N-terminal domain of TDP-43.
Prakash A, Kumar V, Meena NK, Lynn AM. Prakash A, et al. RSC Adv. 2018 May 30;8(35):19835-19845. doi: 10.1039/c8ra03368d. eCollection 2018 May 25. RSC Adv. 2018. PMID: 35548664 Free PMC article. - Monomerization of TDP-43 is a key determinant for inducing TDP-43 pathology in amyotrophic lateral sclerosis.
Oiwa K, Watanabe S, Onodera K, Iguchi Y, Kinoshita Y, Komine O, Sobue A, Okada Y, Katsuno M, Yamanaka K. Oiwa K, et al. Sci Adv. 2023 Aug 4;9(31):eadf6895. doi: 10.1126/sciadv.adf6895. Epub 2023 Aug 4. Sci Adv. 2023. PMID: 37540751 Free PMC article. - Linking hnRNP Function to ALS and FTD Pathology.
Purice MD, Taylor JP. Purice MD, et al. Front Neurosci. 2018 May 15;12:326. doi: 10.3389/fnins.2018.00326. eCollection 2018. Front Neurosci. 2018. PMID: 29867335 Free PMC article. Review. - Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis.
Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK. Prasad A, et al. Front Mol Neurosci. 2019 Feb 14;12:25. doi: 10.3389/fnmol.2019.00025. eCollection 2019. Front Mol Neurosci. 2019. PMID: 30837838 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources