TGF-β1 regulates the expression and transcriptional activity of TAZ protein via a Smad3-independent, myocardin-related transcription factor-mediated mechanism - PubMed (original) (raw)
TGF-β1 regulates the expression and transcriptional activity of TAZ protein via a Smad3-independent, myocardin-related transcription factor-mediated mechanism
Maria Zena Miranda et al. J Biol Chem. 2017.
Abstract
Hippo pathway transcriptional coactivators TAZ and YAP and the TGF-β1 (TGFβ) effector Smad3 regulate a common set of genes, can physically interact, and exhibit multilevel cross-talk regulating cell fate-determining and fibrogenic pathways. However, a key aspect of this cross-talk, TGFβ-mediated regulation of TAZ or YAP expression, remains uncharacterized. Here, we show that TGFβ induces robust TAZ but not YAP protein expression in both mesenchymal and epithelial cells. TAZ levels, and to a lesser extent YAP levels, also increased during experimental kidney fibrosis. Pharmacological or genetic inhibition of Smad3 did not prevent the TGFβ-induced TAZ up-regulation, indicating that this canonical pathway is dispensable. In contrast, inhibition of p38 MAPK, its downstream effector MK2 (e.g. by the clinically approved antifibrotic pirferidone), or Akt suppressed the TGFβ-induced TAZ expression. Moreover, TGFβ elevated TAZ mRNA in a p38-dependent manner. Myocardin-related transcription factor (MRTF) was a central mediator of this effect, as MRTF silencing/inhibition abolished the TGFβ-induced TAZ expression. MRTF overexpression drove the TAZ promoter in a CC(A/T-rich)6GG (CArG) box-dependent manner and induced TAZ protein expression. TGFβ did not act by promoting nuclear MRTF translocation; instead, it triggered p38- and MK2-mediated, Nox4-promoted MRTF phosphorylation and activation. Functionally, higher TAZ levels increased TAZ/TEAD-dependent transcription and primed cells for enhanced TAZ activity upon a second stimulus (i.e. sphingosine 1-phosphate) that induced nuclear TAZ translocation. In conclusion, our results uncover an important aspect of the cross-talk between TGFβ and Hippo signaling, showing that TGFβ induces TAZ via a Smad3-independent, p38- and MRTF-mediated and yet MRTF translocation-independent mechanism.
Keywords: SMAD transcription factor; TAZ; fibrosis; kidney; myocardin-related transcription factor; p38 MAPK; transforming growth factor beta (TGF-B).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
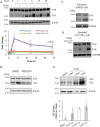
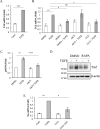
Figure 1.
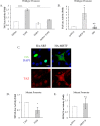
TGFβ induces a rapid increase in TAZ protein expression both in mesenchymal and epithelial cells. TAZ levels also increase in experimental kidney fibrosis. A, C3H/10T1/2 cells were treated with 5 ng/ml TGFβ for the indicated times, and then TAZ and YAP protein levels were determined by Western blotting and densitometry (n = 5). Data are normalized to β-actin as loading control. Similar results were obtained using tubulin or GAPDH as loading controls. B, C3H/10T1/2 cells were treated with vehicle (DMSO) or ALK5 inhibitor SB431542 (10 μ
m
, 30-min preincubation) and then exposed to TGFβ for 6 h. Changes in TAZ and YAP expression were detected as in A. C and D, NRKF (C) and LLC-PK1 kidney tubular cells (D) were incubated without or with TGFβ for 48 h and then processed for Western blotting for the indicated proteins. E, whole-kidney lysates were prepared from mice 1, 3, and 7 days after sham operation or UUO and probed for the indicated proteins. Representative blots are shown from four similar experiments. Densitometric quantitation of TAZ expression normalized to GAPDH is shown below the blot. Errors bars represent standard deviation.
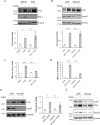
Figure 2.
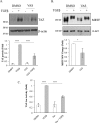
Canonical TGFβ signaling pathway is not critical for TGFβ-induced TAZ expression. A, C3H/10T1/2 cells were treated with vehicle (DMSO) or SIS3 (10 μ
m
, 30-min preincubation) and then exposed to TGFβ for 6 h. TAZ expression was determined by Western blotting and quantified by densitometry (graph below the blot). TAZ expression in the TGFβ-treated samples was taken as unity (n = 3). Membranes were also developed for plasminogen activator inhibitor (PAI-1), a direct transcriptional target of Smad3. B, C3H/10T1/2 cells were transfected with 50 n
m
non-related (siNR) or Smad3-specific siRNA (siSmad3) for 24 h and then, where indicated, exposed to TGFβ for 6 h and processed as in A. TAZ expression (graph below the blot) was quantified by densitometry (n = 3). C and D, to verify the efficiency of SIS3 (C) and the siRNA (D) on Smad3 activity, luciferase assays were performed using an SBE firefly luciferase reporter construct (SBE-Luc). C3H/10T1/2 cells were cotransfected with SBE-Luc and Renilla luciferase (for normalization), 24 h later serum-starved (2 h), and then incubated in the presence or absence of TGFβ for 6 h. E, LLC-PK1 cells were transfected with siNR or siSmad3 (50 n
m
, 24 h), and then exposed to TGFβ for 24 h. TAZ expression was quantified as in A. F, C3H/10T1/2 cells were transfected with siNR or siSmad2 (50 n
m
, 24 h), and then exposed to TGFβ for 6 h. Changes in TAZ expression were determined through Western blotting. n.s., non-significant.
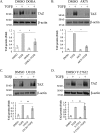
Figure 3.
Non-canonical signaling through p38 and Akt plays essential roles in the mediation of TGFβ-induced TAZ expression, whereas the activation of ERK and Rho kinase is dispensable. Cells were pretreated with vehicle (DMSO) or the corresponding pharmacological inhibitor (10 μ
m
) for 30 min and, where indicated, exposed to TGFβ for 6 h. TAZ expression was quantified by densitometry using the TGFβ-treated sample as unity. The inhibitors used were as follows: A, DORA for p38 (n = 4); B, AKTi for Akt1/2 (n = 3); C, U0126 for MEK/ERK1/2 (n = 7); and D, Y27632 for ROK (n = 10). ns, non-significant.
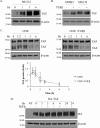
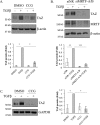
Figure 4.
Altered TAZ degradation cannot account for the TGFβ-induced increase in TAZ expression. A, to assess the role of the proteasome in regulating steady-state TAZ levels, C3H/10T1/2 cells were treated with the proteasome inhibitor MG132 (40 μ
m
) for the indicated times, and changes in TAZ expression were determined by Western blotting. B and C, to assess whether TGFβ prevents the constitutive degradation of TAZ, cells were treated with MG132 (B) or the protein synthesis inhibitor CHX (25 μg/ml) (C) in the absence or presence of TGFβ. B, cells were treated with vehicle (DMSO) or MG132 (40 μ
m
) and, where indicated, exposed to TGFβ for 6 h. C, cell lysates were collected at the allotted times and change in TAZ protein was quantified relative to the 0-h conditions. The graph shows the respective degradation profiles for both conditions (n = 3). D, cells were transfected with a HA-TAZ construct (0.5 μg/ml) and 24 h later treated with or without TGFβ for the indicated times. HA-TAZ expression was monitored using an anti-HA antibody.
Figure 5.
TGFβ increases TAZ mRNA via a p38- and SRF/MRTF inhibitor-sensitive mechanism. A, C3H/10T1/2 cells were left untreated or stimulated with TGFβ for 6 h. qPCR was performed, and TAZ mRNA was normalized to GAPDH mRNA. Fold change was calculated using the Pfaffl method. Cntrl, control. B, C3H/10T1/2 cells were pretreated with vehicle (DMSO) or pharmacological inhibitors DORA (10 μ
m
), AKTi (10 μ
m
), or CCG (3 μ
m
) for 30 min and then exposed to TGFβ for 3 h. qPCR was performed, and TAZ mRNA was normalized to an alternative housekeeping gene, RLP13a mRNA. C, immunofluorescence quantitation of S6 protein phosphorylation. Cells were pretreated with vehicle (DMSO) or AKTi (10 μ
m
, AKTi1/2) for 30 min and, where indicated, exposed to TGFβ for 6 h. S6 phosphorylation was determined by immunofluorescence staining and automated image analysis as described under “Experimental procedures” (n = 3 experiments in each of which 1000–3000 cells/condition were quantified). D, cells were pretreated with vehicle (DMSO) or rapamycin (100 n
m
, RAPA) for 30 min and, where indicated, exposed to TGFβ for 6 h. Changes in TAZ expression was determined by Western blotting. E, LLC-PK1 cells were pretreated with DMSO or CCG, then exposed to TGFβ for 24 h, and processed for qPCR to determine TAZ mRNA as in A (n = 3). ns, non-significant.
Figure 6.
MRTF is necessary for TGFβ-induced TAZ expression. A and C, C3H/10T1/2 cells (A) or LLC-PK1 cells (C) were pretreated with vehicle (DMSO) or the MRTF inhibitor CCG (3 μ
m
) and, where indicated, exposed to TGFβ for 6 or 24 h, respectively (n = 6). B, C3H/10T1/2 cells were treated with NR or MRTF-A/B siRNA (100 and 200 n
m
for MRTF-A and -B, respectively) for 48 h (n = 3). TAZ expression was then quantified by densitometry with the TGFβ-treated sample taken as unity.
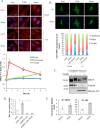
Figure 7.
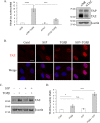
TGFβ does not enhance the nuclear localization of MRTF but induces enhanced MRTF/SRF activity. A, C3H/10T1/2 cells were left untreated or stimulated with TGFβ or 10% fetal bovine serum (FBS) for the indicated times. Cells were then stained for endogenous MRTF and the nuclear marker DAPI, and MRTF distribution was visualized by immunofluorescence microscopy. The graph shows fold changes in the nuclear-to-cytoplasmic ratio of MRTF, determined by the Metamorph software (see “Experimental procedures”) and normalized for each condition to the untreated control (Cntrl) at 0 h. For each time point and treatment, the ratios were determined in 22–30 cells. Scale bar, 20 μm. B, C3H/10T1/2 cells were transfected with GFP-MRTF and subsequently either left untreated or stimulated with TGFβ or 10% FBS. The graph shows the quantification of subcellular distribution (nuclear, cytoplasmic, or diffuse) of GFP-MRTF, using >100 cells in each category at each time point. Scale bar, 20 μm. C, MRTF localization was determined using nuclear preparations. C3H/10T1/2 cells were incubated without or with TGFβ for 3 h. Cytoplasmic and nuclear fractions were then separated (see under “Experimental procedures”) and probed for MRTF, tubulin as a cytosolic marker, and histone H3 as a nuclear marker. D, to determine the effect of TGFβ on MRTF activity, luciferase assays were performed using a CArG box containing firefly luciferase reporter construct (3DA). Cells were cotransfected with 3DA and Renilla luciferase along with NR or MRTF-A/B siRNA (100/200 n
m
, respectively, 48 h). Cells were then incubated in the presence or absence of TGFβ for 6 h, and 3DA-Luc activity was measured and normalized to that of Renilla. E, for chromatin immunoprecipitation, C3H/10T1/2 cells were treated with vehicle (DMSO) or TGFβ for 3 h. Chromatin–protein complexes were then cross-linked, sheared, and immunoprecipitated (IP) using either anti-MRTF or anti-SRF antibodies as described under “Experimental procedures.” qPCR was performed to quantify the coprecipitating TAZ promoter, using specific primers flanking the CArG box. Data are normalized to the signal obtained by control (unrelated) IgG.
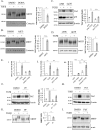
Figure 8.
TGFβ induces post-translational modification of MRTF in a p38- and MK2-dependent manner. A and B, C3H/10T1/2 cells were pretreated with vehicle (DMSO) or DORA (A) or AKTi (B) and, where indicated, exposed to TGFβ for 6 h. Whole-cell extracts were run in 6% SDS gels, and the relative shift is the molecular mass of MRTF (see dotted lines) was determined and normalized to the change detected in the vehicle-treated controls as described under “Experimental procedures.” C and D, cells were transfected with 300 n
m
non-related (siNR) or α (100 n
m
)-, β (100 n
m
)-, and γ (100 n
m
)-p38 siRNA (sip38) for 48 h and then, where indicated, exposed to TGFβ for 6 h. Cortactin was used as loading control. TAZ expression (C) and the relative molecular shift of MRTF (D) were assessed by Western blotting (n = 3) as in Figs. 1_A_ and 8_B_, respectively. α-Actinin (α-act) was used as loading control. E, cells were cotransfected with 3DA firefly reporter and Renilla luciferase and, where indicated (panel iii) with Smad3 siRNA or NR siRNA for 24 h. Subsequently cells were preincubated with vehicle (DMSO) or DORA (panel i) or AKTi (panel ii) for 30 min. Cells were then exposed to TGFβ for 6 h, as indicated. Normalized luciferase activities were determined (n = 3 for each condition). F and G, C3H/10T1/2 cells were pretreated with vehicle or the MK2 inhibitor PF-3644022 (PF, 10 μ
m
) followed by 6 h of treatment with TGFβ as indicated and then processed for Western blotting to assess TAZ expression (F) or the shift of MRTF (G) (n = 3). H and I, conditions were as in F and G, except the cells were pretreated where indicated with the antifibrotic drug, pirfenidone (Pirf, 1 mg/ml). n.s., non-significant.
Figure 9.
TGFβ or overexpression of MRTF but not SRF can induce TAZ protein expression and TAZ promoter activation. A, C3H/10T1/2 cells were cotransfected with a ≈ 1.2 kb segment of the TAZ promoter coupled to firefly luciferase (TAZ-Luc) along with Renilla luciferase for 24 h. Subsequently cells were pretreated with vehicle (DMSO) or DORA (10 μ
m
) for 30 min, then stimulated with TGFβ for 24 h. Normalized TAZ promoter activity was determined by the dual-luciferase assay. B, to assess whether MRTF or SRF was sufficient to drive the TAZ promoter, C3H/10T1/2 cells were cotransfected with TAZ-Luc and Renilla along with either FLAG-MRTF or HA-SRF. Twenty four h later TAZ-Luc activity was measured. C, verification of expression of FLAG-MRTF and HA-SRF and assessment of their impact on TAZ expression was achieved by double immunofluorescence staining for the corresponding epitope tag and endogenous TAZ. Note that HA-SRF is strongly expressed and exhibits robust nuclear localization, yet it does not cause a detectable increase in TAZ expression, as opposed to FLAG-MRTF. Scale bar, 20 μm. D, C3H/10T1/2 cells were transfected with a mutant version (TAZmut-Luc) of TAZ-Luc, in which the CArG box has been inactivated, and with Renilla. Twenty four h later cells were left untreated or treated with TGFβ for an additional 24 h, and luciferase activities were determined (n = 3). E, conditions were as in D, except cells were transfected with FLAG-MRTF-B along with the TAZmut-Luc/Renilla system for 24 h, and luciferase activities were measured without further treatment (n = 3). Cntrl, control. n.s., non-significant.
Figure 10.
Nox4 contributes to TGFβ-induced TAZ expression and the post-translational modification of MRTF. A and B, cells were pretreated with vehicle (DMSO) or VAS2870 (10 μ
m
, VAS), a potent inhibitor of Nox4 for 30 min and, where indicated, exposed to TGFβ for 6 h. TAZ expression (A) and the shift in the molecular mass of MRTF (B) were then quantified (n = 3). C, C3H/10T1/2 cells were transfected with TAZ-Luc and Renilla luciferase for 24 h, followed by pretreatment with DMSO or VAS2870 and, where indicated, with TGFβ for 24 h (n = 3).
Figure 11.
TGFβ-induced TAZ expression increases TAZ-dependent transcriptional activity and primes cells for augmented TAZ activity in response to a stimulus inducing TAZ translocation. A, to demonstrate TAZ-dependent transcriptional activity, luciferase assays were performed using a reporter construct harboring tandem TEAD-binding elements in front of firefly luciferase (TEAD-Luc). Cells were transfected with the TEAD-Luc/Renilla system along with siNR or TAZ-specific siRNA (siTAZ, 25 n
m
) for 24 h and then left untreated or exposed to TGFβ for an additional 24 h, followed by the determination of TEAD activity (n = 3). TAZ knockdown was confirmed by Western blotting performed in parallel. B, cells were incubated in the absence or presence of TGFβ for 5 h and subsequently challenged, where indicated, with S1P (1 μ
m
) for 1 h. Cells were then stained for endogenous TAZ, and nuclei were visualized by DAPI. Scale bar, 20 μm. C and D, to demonstrate the priming effect of TGFβ on TAZ activity, cells were pretreated with TGFβ for 6 h and then further stimulated with S1P (1 μ
m
) to induce translocation of TAZ to the nucleus. The effect on TAZ protein expression (C) and TEAD-Luc activity (D) was then measured (n = 3). Cntrl, control. n.s., non-significant.
Similar articles
- Myocardin-related Transcription Factor Regulates Nox4 Protein Expression: LINKING CYTOSKELETAL ORGANIZATION TO REDOX STATE.
Rozycki M, Bialik JF, Speight P, Dan Q, Knudsen TE, Szeto SG, Yuen DA, Szászi K, Pedersen SF, Kapus A. Rozycki M, et al. J Biol Chem. 2016 Jan 1;291(1):227-43. doi: 10.1074/jbc.M115.674606. Epub 2015 Nov 10. J Biol Chem. 2016. PMID: 26555261 Free PMC article. - YAP/TAZ regulates TGF-β/Smad3 signaling by induction of Smad7 via AP-1 in human skin dermal fibroblasts.
Qin Z, Xia W, Fisher GJ, Voorhees JJ, Quan T. Qin Z, et al. Cell Commun Signal. 2018 Apr 25;16(1):18. doi: 10.1186/s12964-018-0232-3. Cell Commun Signal. 2018. PMID: 29695252 Free PMC article. - Inhibitors of oxygen sensing prolyl hydroxylases regulate nuclear localization of the transcription factors Smad2 and YAP/TAZ involved in CTGF synthesis.
Preisser F, Giehl K, Rehm M, Goppelt-Struebe M. Preisser F, et al. Biochim Biophys Acta. 2016 Aug;1863(8):2027-36. doi: 10.1016/j.bbamcr.2016.05.001. Epub 2016 May 5. Biochim Biophys Acta. 2016. PMID: 27155083 - Reciprocal regulation of YAP/TAZ by the Hippo pathway and the Small GTPase pathway.
Jang JW, Kim MK, Bae SC. Jang JW, et al. Small GTPases. 2020 Jul;11(4):280-288. doi: 10.1080/21541248.2018.1435986. Epub 2018 Apr 20. Small GTPases. 2020. PMID: 29457552 Free PMC article. Review. - [Regulation of differentiation of mesenchymal stem cells by the Hippo pathway effectors TAZ/YAP].
Men T, Piao SH, Teng CB. Men T, et al. Yi Chuan. 2013 Nov;35(11):1283-90. doi: 10.3724/sp.j.1005.2013.01283. Yi Chuan. 2013. PMID: 24579311 Review. Chinese.
Cited by
- YAP/TAZ Signaling as a Molecular Link between Fibrosis and Cancer.
Noguchi S, Saito A, Nagase T. Noguchi S, et al. Int J Mol Sci. 2018 Nov 20;19(11):3674. doi: 10.3390/ijms19113674. Int J Mol Sci. 2018. PMID: 30463366 Free PMC article. Review. - Profibrotic epithelial phenotype: a central role for MRTF and TAZ.
Bialik JF, Ding M, Speight P, Dan Q, Miranda MZ, Di Ciano-Oliveira C, Kofler MM, Rotstein OD, Pedersen SF, Szászi K, Kapus A. Bialik JF, et al. Sci Rep. 2019 Mar 13;9(1):4323. doi: 10.1038/s41598-019-40764-7. Sci Rep. 2019. PMID: 30867502 Free PMC article. - Recent Advances in Understandings Towards Pathogenesis and Treatment for Intrauterine Adhesion and Disruptive Insights from Single-Cell Analysis.
Leung RK, Lin Y, Liu Y. Leung RK, et al. Reprod Sci. 2021 Jul;28(7):1812-1826. doi: 10.1007/s43032-020-00343-y. Epub 2020 Oct 30. Reprod Sci. 2021. PMID: 33125685 Free PMC article. Review. - MRTF-A-NF-κB/p65 axis-mediated PDL1 transcription and expression contributes to immune evasion of non-small-cell lung cancer via TGF-β.
Du F, Qi X, Zhang A, Sui F, Wang X, Proud CG, Lin C, Fan X, Li J. Du F, et al. Exp Mol Med. 2021 Sep;53(9):1366-1378. doi: 10.1038/s12276-021-00670-3. Epub 2021 Sep 21. Exp Mol Med. 2021. PMID: 34548615 Free PMC article. - Nucleocytoplasmic Shuttling of the Mechanosensitive Transcription Factors MRTF and YAP /TAZ.
Kofler M, Kapus A. Kofler M, et al. Methods Mol Biol. 2021;2299:197-216. doi: 10.1007/978-1-0716-1382-5_15. Methods Mol Biol. 2021. PMID: 34028745
References
- Piccolo S., Dupont S., and Cordenonsi M. (2014) The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 - PubMed
- Low B. C., Pan C. Q., Shivashankar G. V., Bershadsky A., Sudol M., and Sheetz M. (2014) YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 588, 2663–2670 - PubMed
- Dupont S. (2016) Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res. 343, 42–53 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials