Permissivity of Dipeptidyl Peptidase 4 Orthologs to Middle East Respiratory Syndrome Coronavirus Is Governed by Glycosylation and Other Complex Determinants - PubMed (original) (raw)
Permissivity of Dipeptidyl Peptidase 4 Orthologs to Middle East Respiratory Syndrome Coronavirus Is Governed by Glycosylation and Other Complex Determinants
Kayla M Peck et al. J Virol. 2017.
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) utilizes dipeptidyl peptidase 4 (DPP4) as an entry receptor. While bat, camel, and human DPP4 support MERS-CoV infection, several DPP4 orthologs, including mouse, ferret, hamster, and guinea pig DPP4, do not. Previous work revealed that glycosylation of mouse DPP4 plays a role in blocking MERS-CoV infection. Here, we tested whether glycosylation also acts as a determinant of permissivity for ferret, hamster, and guinea pig DPP4. We found that, while glycosylation plays an important role in these orthologs, additional sequence and structural determinants impact their ability to act as functional receptors for MERS-CoV. These results provide insight into DPP4 species-specific differences impacting MERS-CoV host range and better inform our understanding of virus-receptor interactions associated with disease emergence and host susceptibility.IMPORTANCE MERS-CoV is a recently emerged zoonotic virus that is still circulating in the human population with an ∼35% mortality rate. With no available vaccines or therapeutics, the study of MERS-CoV pathogenesis is crucial for its control and prevention. However, in vivo studies are limited because MERS-CoV cannot infect wild-type mice due to incompatibilities between the virus spike and the mouse host cell receptor, mouse DPP4 (mDPP4). Specifically, mDPP4 has a nonconserved glycosylation site that acts as a barrier to MERS-CoV infection. Thus, one mouse model strategy has been to modify the mouse genome to remove this glycosylation site. Here, we investigated whether glycosylation acts as a barrier to infection for other nonpermissive small-animal species, namely, ferret, guinea pig, and hamster. Understanding the virus-receptor interactions for these DPP4 orthologs will help in the development of additional animal models while also revealing species-specific differences impacting MERS-CoV host range.
Keywords: DPP4; MERS-coronavirus; animal models; glycosylation; host range; host range expansion; orthologs.
Copyright © 2017 American Society for Microbiology.
Figures
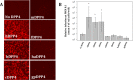
FIG 1
Permissivity of DPP4 orthologs to MERS-CoV. (A) Seven DPP4 orthologs were tested for their ability to support infection by rMERS-CoV-RFP. DPP4 constructs were transfected into HEK 293T cells and infected at an MOI of 5 at ∼20 h posttransfection. Cells were imaged for fluorescence at ∼24 hpi. hDPP4, human DPP4; cDPP4, camel DPP4; bDPP4, bat DPP4; mDPP4, mouse DPP4; fDPP4, ferret DPP4; haDPP4, hamster DPP4; gpDPP4, guinea pig DPP4. (B) Mean fluorescent cell counts of MERS-CoV infection utilizing various DPP4 orthologs. Cells were infected at an MOI of 0.1 and the numbers of infected cells counted at ∼72 hpi. Each DPP4 ortholog was measured in triplicate. Only hDPP4, bDPP4, and cDPP4 had levels of infection significantly higher than those seen in the absence of DPP4 (*, P < 0.05 [Student's t test]). All DPP4 orthologs had significantly lower levels of infection than hDPP4 (P < 0.05 [Student's t test]). The levels of infection seen between bDPP4 and cDPP4 were not significantly different. Error bars indicate mean values ± 1 standard deviation.
FIG 2
Sequence and structural comparison of nonpermissive DPP4 orthologs. (A) Structural comparison of threaded molecules (30) of mDPP4 (orange), fDPP4 (green), haDPP4 (blue), and gpDPP4 (purple) overlaid on hDPP4 (yellow) complexed with the MERS-CoV RBD (red) (PDB code
4L72
). (B) Sequence alignment of permissive (human, camel, bat; blue) and nonpermissive (mouse, ferret, hamster, guinea pig; red) DPP4 amino acid sequences. Residue 330 is numbered relative to mDPP4. Boxes represent glycosylation sites that are either unique to nonpermissive species (black) or shared with a permissive species (gray). (C) hDPP4 (yellow) complexed with the MERS-CoV RBD (red) (PDB code
4L72
). Residues aligning to the ferret (green), hamster and mouse (blue), and guinea pig (purple) glycosylation sites are highlighted. Dashed-line circles indicate the regions of the DPP4 molecule that correspond to blades IV and V.
FIG 3
DPP4 ortholog glycosylation knockout mutants. (A) Neither wild-type nor glycosylation knockout (−gly) DPP4 molecules for ferret (fDPP4), hamster (haDPP4), or guinea pig (gpDPP4) support infection by MERS-CoV. (B) Successful removal of glycosylation is supported by an ∼2.5-kDa downward shift seen via Western blotting. The top blot represents DPP4, and the bottom blot represents β-actin as a control. (C) Fluorescent cell counts from MERS-CoV infection utilizing various DPP4 orthologs and their respective glycosylation knockout mutants. Cells were infected at an MOI of 0.1, and numbers of infected cells were counted at 72 hpi. The level of each DPP4 ortholog was measured in triplicate. Only hDPP4 had levels of infection significantly higher than those seen in the absence of DPP4 (*, P < 0.05 [Student's t test]). The remaining DPP4 orthologs and glycosylation knockouts had infection levels that were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
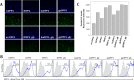
FIG 4
DPP4 and mutant variants are expressed on the surface of cells as evidenced by results of immunofluorescence assay (A) and flow cytometry (B and C). (A) Cells were transfected with each DPP4 ortholog, fixed, and probed with primary goat anti-DPP4 polyclonal antibody (R&D Systems) at 1:50 and secondary donkey anti-goat Alexa Fluor 488 (Life Technologies) at 1:500. Cells were imaged at a magnification of ×20 for DAPI (300 ms exposure) and DPP4 (1.5 s exposure). (B) DPP4 expression frequencies (blue outlined histogram) by DPP4 construct after subtraction of background from replicate wells stained with secondary donkey anti-goat IgG (H+L) Alexa Fluor 488 antibody only (gray-shaded histogram). Percentage values represent averages of results across two duplicate wells. Max, maximum. (C) Geometric mean fluorescence intensity of the DPP4-positive populations for each DPP4 construct.
FIG 5
Many amino acid changes are required to make fDPP4 and haDPP4 permissive to MERS-CoV infection. (A) Removing glycosylation alone did not confer permissivity to haDPP4. However, combining three amino acid changes on blade V (starting at residue 289) with the glycosylation knockout mutant on blade IV (N332A) resulted in high levels of MERS-CoV infection. Sequences show the alignment between hDPP4 and haDPP4, with the black boxes indicating the amino acids that were swapped from hDPP4 into haDPP4. (B) Removing glycosylation alone did not confer permissivity to fDPP4. However, introducing a set of amino acid changes on blade V (starting at residue 278) and blade IV (starting at residue 330) allowed fDPP4 to support MERS-CoV infection [fDPP4 (278) (330)]. Sequences show the alignment between hDPP4 and fDPP4, with the black boxes indicating the amino acids that were swapped from hDPP4 into fDPP4. Note that fDPP4 −gly is a negative control and includes only the single point mutation N331A. (C) Western blot analysis of fDPP4 and haDPP4 and designated variants for DPP4 and β-actin expression. Successful glycosylation knockout is indicated by a downward shift of ∼2.5 kDa. (D) Fluorescent cell counts of MERS-CoV infection utilizing DPP4 orthologs. Cells were infected at an MOI of 0.1, and numbers of red cells were counted at 72 hpi. Each DPP4 ortholog was measured in triplicate. hDPP4, fDPP4 (278) (330), and haDPP4 (289), −gly had levels of infection that were significantly greater than those seen in the absence of DPP4 (P < 0.05 [Student's t test]). fDPP4 −gly and haDPP4 −gly infection levels were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
FIG 6
Bat and guinea pig DPP4 share the same glycosylation site downstream of the site identified to be important in mDPP4 (Fig. 2B). (A) Removal of the glycosylation site from bDPP4 showed no decrease in infection, while removal of glycosylation from gpDPP4 resulted in no increase in infection. (B) Western blot analysis of bDPP4 and gpDPP4 and their respective glycosylation knockout mutants for DPP4 and β-actin expression. Successful glycosylation knockout is indicated by a downward shift of ∼2.5 kDa. (C) DPP4 and mutant variants are expressed on the surface of cells, visible by immunofluorescence. Cells were transfected with each DPP4 ortholog, fixed, and probed with primary goat anti-DPP4 polyclonal antibody (R&D Systems) at 1:50 and secondary donkey anti-goat Alexa Fluor 488 (Life Technologies) at 1:500. Cells were imaged at a magnification of ×20 for DAPI (300-ms exposure) and DPP4 (1.5-s exposure). (D) Fluorescent cell counts of MERS-CoV infection utilizing various DPP4 orthologs. Cells were infected at an MOI of 0.1, and numbers of infected cells were counted at 72 hpi. Each DPP4 ortholog was measured in triplicate. hDPP4, bDPP4, and bDPP4 −gly had levels of infection that are significantly higher than those seen in the absence of DPP4 (P < 0.05 [Student's t test]). gpDPP4 and gpDPP4 −gly infection levels were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
FIG 7
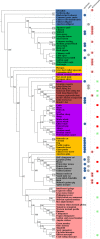
DPP4 protein phylogenetic tree based on amino acid sequences. Shaded colors indicate the group each species falls in. Blue, reptiles and amphibians; green, avian species; orange, other mammals; red, Chiroptera (bats); purple, ungulates; gray, rodents; pink: primates. Colored circles to the right of the species names indicate whether the sequence has a glycosylation site upstream of (first column), at the same site as (second column), or downstream of (third column) the NXT glycosylation site in mDPP4 (residues 332 to 334). Numbers inside the circle designate how many amino acids upstream (or downstream) the N of the NXT or NXS glycosylation site is. For the second column, a circle indicates that there was a glycosylation site aligning to the site present in mDPP4. Squares in the rightmost column indicate permissive (green) or nonpermissive (red) species, as determined from either in vivo or in vitro studies. Numbers indicate bootstrap support values >50.
Similar articles
- Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection.
Peck KM, Cockrell AS, Yount BL, Scobey T, Baric RS, Heise MT. Peck KM, et al. J Virol. 2015 Apr;89(8):4696-9. doi: 10.1128/JVI.03445-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653445 Free PMC article. - Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4.
van Doremalen N, Miazgowicz KL, Milne-Price S, Bushmaker T, Robertson S, Scott D, Kinne J, McLellan JS, Zhu J, Munster VJ. van Doremalen N, et al. J Virol. 2014 Aug;88(16):9220-32. doi: 10.1128/JVI.00676-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899185 Free PMC article. - CD8+ T Cells and Macrophages Regulate Pathogenesis in a Mouse Model of Middle East Respiratory Syndrome.
Coleman CM, Sisk JM, Halasz G, Zhong J, Beck SE, Matthews KL, Venkataraman T, Rajagopalan S, Kyratsous CA, Frieman MB. Coleman CM, et al. J Virol. 2016 Dec 16;91(1):e01825-16. doi: 10.1128/JVI.01825-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795435 Free PMC article. - Coronavirus Host Range Expansion and Middle East Respiratory Syndrome Coronavirus Emergence: Biochemical Mechanisms and Evolutionary Perspectives.
Peck KM, Burch CL, Heise MT, Baric RS. Peck KM, et al. Annu Rev Virol. 2015 Nov;2(1):95-117. doi: 10.1146/annurev-virology-100114-055029. Epub 2015 Aug 7. Annu Rev Virol. 2015. PMID: 26958908 Review. - Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Infection, Immunological Response, and Vaccine Development.
Mubarak A, Alturaiki W, Hemida MG. Mubarak A, et al. J Immunol Res. 2019 Apr 7;2019:6491738. doi: 10.1155/2019/6491738. eCollection 2019. J Immunol Res. 2019. PMID: 31089478 Free PMC article. Review.
Cited by
- Why COVID-19 Transmission Is More Efficient and Aggressive Than Viral Transmission in Previous Coronavirus Epidemics?
Elrashdy F, Redwan EM, Uversky VN. Elrashdy F, et al. Biomolecules. 2020 Sep 11;10(9):1312. doi: 10.3390/biom10091312. Biomolecules. 2020. PMID: 32933047 Free PMC article. Review. - Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
Yuan Y, Qi J, Peng R, Li C, Lu G, Yan J, Wang Q, Gao GF. Yuan Y, et al. J Virol. 2020 Feb 14;94(5):e01387-19. doi: 10.1128/JVI.01387-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31776269 Free PMC article. - Current understanding of middle east respiratory syndrome coronavirus infection in human and animal models.
Wang Y, Sun J, Zhu A, Zhao J, Zhao J. Wang Y, et al. J Thorac Dis. 2018 Jul;10(Suppl 19):S2260-S2271. doi: 10.21037/jtd.2018.03.80. J Thorac Dis. 2018. PMID: 30116605 Free PMC article. Review. - Origin and evolution of pathogenic coronaviruses.
Cui J, Li F, Shi ZL. Cui J, et al. Nat Rev Microbiol. 2019 Mar;17(3):181-192. doi: 10.1038/s41579-018-0118-9. Nat Rev Microbiol. 2019. PMID: 30531947 Free PMC article. Review. - Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells.
Chen YN, Hsu HC, Wang SW, Lien HC, Lu HT, Peng SK. Chen YN, et al. Pathogens. 2019 Nov 22;8(4):259. doi: 10.3390/pathogens8040259. Pathogens. 2019. PMID: 31766704 Free PMC article.
References
- Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, Muller MA, Annan A, Vallo P, Adu-Sarkodie Y, Kruppa TF, Drosten C. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis 15:1377–1384. doi:10.3201/eid1509.090224. - DOI - PMC - PubMed
- Menachery VD, Yount BL, Debbink K, Agnihothram SS, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. doi:10.1038/nm.3985. - DOI - PMC - PubMed
- Menachery VD, Yount BL, Sims AC, Debbink K, Agnihothram SS, Gralinksi LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113:3048–3053. doi:10.1073/pnas.1517719113. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous