Vestigial-like 2 contributes to normal muscle fiber type distribution in mice - PubMed (original) (raw)
Vestigial-like 2 contributes to normal muscle fiber type distribution in mice
Masahiko Honda et al. Sci Rep. 2017.
Abstract
Skeletal muscle is composed of heterogeneous populations of myofibers that are classified as slow- and fast-twitch fibers. The muscle fiber-type is regulated in a coordinated fashion by multiple genes, including transcriptional factors and microRNAs (miRNAs). However, players involved in this regulation are not fully elucidated. One of the members of the Vestigial-like factors, Vgll2, is thought to play a pivotal role in TEA domain (TEAD) transcription factor-mediated muscle-specific gene expression because of its restricted expression in skeletal muscles of adult mice. Here, we generated Vgll2 null mice and investigated Vgll2 function in adult skeletal muscles. These mice presented an increased number of fast-twitch type IIb fibers and exhibited a down-regulation of slow type I myosin heavy chain (MyHC) gene, Myh7, which resulted in exercise intolerance. In accordance with the decrease in Myh7, down-regulation of miR-208b, encoded within Myh7 gene and up-regulation of targets of miR-208b, Sox6, Sp3, and Purβ, were observed in Vgll2 deficient mice. Moreover, we detected the physical interaction between Vgll2 and TEAD1/4 in neonatal skeletal muscles. These results suggest that Vgll2 may be both directly and indirectly involved in the programing of slow muscle fibers through the formation of the Vgll2-TEAD complex.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Figure 1
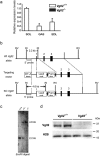
Expression patterns of Vgll2 mRNA expression in several muscles, and generation of Vgll2-deficient mice. (a) Vgll2 mRNA levels were measured by qPCR in the soleus (SOL), gastrocnemius (GAS), and extensor digitorum longus (EDL) muscles from 12-week-old Vgll2 +/+ and Vgll2 −/− mice (n = 6). *P < 0.05 vs. Vgll2 +/+ soleus muscles. Data are presented as mean ± SEM. For comparison, the expression level in Vgll2 +/+ soleus was arbitrarily set at 1. (b) Schematic diagram showing the strategy used to generate the Vgll2 null allele. The top row depicts the wild-type Vgll2 allele, which consists of three exons (solid boxes). The initiation Met codon for Vgll2 exists in exon 1. The second row depicts the targeting construct. The third row depicts the mutated Vgll2 allele. The lacZ expression cassette with a SV40 polyadenylation signal, which was followed by the loxP-flanked puromycin resistant gene expression cassette (Puro) in the reverse orientation, was fused to the initiation codon of Vgll2. The diphtheria toxin A expression cassette (DT-A) was included at the 5′-end of the targeting vector. The external probe used for Southern blot analysis lies outside the 5′-homologous arm (gray box). RI, _Eco_RI; RV, _Eco_RV. (c) Southern blot analysis for genotyping wild-type (Vgll2 +/+), heterozygous Vgll2-deficient (Vgll2 +/−), and homozygous Vgll2-deficient (Vgll2 −/−) animals. Digestion with _Eco_RI of Vgll2 +/+ and Vgll2 −/− alleles gave rise to fragments of 2.2 and 4.6 kb, respectively. An uncropped original image is shown in Supplementary information. (d) Vgll2 protein levels were assessed by Western blotting using nuclear protein extracts obtained from the soleus muscle of 12-week-old Vgll2 +/+ and Vgll2 −/− mice. Histone H2B served as a loading control. Analyses were performed on four mice per genotype. Data from two mice per genotype are shown. Uncropped original images are shown in Supplementary information.
Figure 2
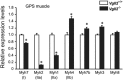
Expression analysis of myosin heavy chain isoforms in neonatal skeletal muscle. Expression levels of MyHC isoforms were measured by qPCR in the gastrocnemius-plantaris-soleus (GPS) muscle complex at postnatal day 7 (P7) of Vgll2 +/+ and Vgll2 −/− mice (n = 8). Myh7 (encoding MyHCI), Myh2 (MyHCIIa), Myh1 (MyHCIIx), Myh4 (MyHCIIb), Myh7b (slow-tonic MyHC), Myh3 (embryonic fast isoform, MyHC-emb), and Myh8 (perinatal fast isoform, MyHC-pn) expression levels were examined. For comparison, the expression level of these genes in Vgll2 +/+ mice was arbitrarily set at 1. Data are presented as mean ± SEM. *P < 0.05 vs. Vgll2 +/+ muscles.
Figure 3
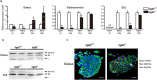
Fiber type composition analysis based on the expression of myosin heavy chain isoforms in adult skeletal muscles. (a) Expression levels of genes encoding MyHC isoforms, Myh7 (I), Myh2 (IIa), Myh1 (IIx), and Myh4 (IIb) were measured by qPCR in the soleus, gastrocnemius, and EDL muscles from 12-week-old Vgll2 +/+ and Vgll2 −/− mice (n = 6). For comparison, the expression level of these genes in Vgll2 +/+ mice was arbitrarily set at 1. Data are presented as mean ± SEM. *P < 0.05 vs. Vgll2 +/+ in each muscle. (b) High resolution gel electrophoresis for MyHC isoform separation of protein extracts isolated from the soleus and EDL muscles of 12-week-old Vgll2 +/+ and Vgll2 −/− mice. Equal amounts (50 ng) of total protein were separated on an 8% acrylamide gel containing glycerol. The gel was then stained with the silver stain method. Uncropped original images are shown in Supplementary information. (c) Representative images of immunostained soleus muscles isolated from 12-week-old Vgll2 +/+ and Vgll2 −/− mice (n = 3). Scale bar: 200 μm.
Figure 4
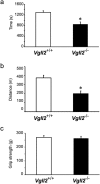
Analysis of muscle functions in Vgll2 null mice. (a,b) Physical endurance was measured by using a treadmill running test. After acclimation, mice ran on a 10% slope with a protocol using increasing speed until mice were exhausted. Exhaustion was defined as the inability of the mice to remain on the treadmill despite physical prodding. Running time (a) and speed were measured, and the distance (b) was calculated. (c) Skeletal muscle strength was assessed by using the grip strength test. Grip strength was measured in each mouse 20 times and the highest value of each experiment was used. Male mice (11–13-week-old) were used in all experiments (n = 8). Data are presented as mean ± SEM. *P < 0.05 vs. Vgll2 +/+ mice.
Figure 5
Physical interaction analysis between endogenous Vgll2 and TEAD1 in skeletal muscles at the neonatal stage. (a) Immunoprecipitation was conducted with the anti-Vgll2 antibody followed by Western blot analysis to detect TEAD1, TEAD4 and Vgll2 in GPS muscle extracts from Vgll2 +/+ and Vgll2 −/− mice at P7. (b) Immunoprecipitation was conducted with an anti-TEAD1 antibody followed by Western blot analysis to detect Vgll2 and TEAD1 in P7 GPS muscle extracts. (c) Immunoprecipitation was conducted with an anti-TEAD4 antibody followed by Western blot analysis to detect Vgll2 and TEAD4 in P7 GPS muscle extracts. Control samples were GPS muscle lysates incubated without anti-Vgll2 or -TEAD1 antibody (no-Ab). Uncropped original images are shown in Supplementary information.
Figure 6
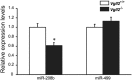
Expression analysis of miRNAs in the neonatal skeletal muscle. miR-208b and miR-499 expression levels in the GPS muscle from Vgll2 +/+ and Vgll2 −/− mice at P7 (n = 8). For comparison, the expression level of these miRNAs in Vgll2 +/+ mice was arbitrarily set at 1. Data are presented as mean ± SEM. *P < 0.05 vs. Vgll2 +/+ muscles.
Figure 7
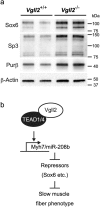
Expression analysis of transcriptional repressor proteins in the neonatal skeletal muscles. (a) Sox6, Sp3, and Purβ protein levels were assessed by Western blotting in the GPS muscle from Vgll2 +/+ and Vgll2 −/− mice at P7. β-Actin was used as a loading control. Analyses were performed on four mice per genotype. Data from two mice per genotype are shown. Uncropped original images are shown in Supplementary information. (b) Schematic representation of the possible mechanism of TEAD1-mediated Vgll2 functions in muscle fiber type regulation.
Similar articles
- MicroRNA-139-5p suppresses myosin heavy chain I and IIa expression via inhibition of the calcineurin/NFAT signaling pathway.
Xu M, Chen X, Huang Z, Chen D, Yu B, Chen H, He J, Zheng P, Luo J, Yu J, Luo Y. Xu M, et al. Biochem Biophys Res Commun. 2018 Jun 12;500(4):930-936. doi: 10.1016/j.bbrc.2018.04.202. Epub 2018 Apr 30. Biochem Biophys Res Commun. 2018. PMID: 29705696 - Transcriptional cofactor Vgll2 is required for functional adaptations of skeletal muscle induced by chronic overload.
Honda M, Tsuchimochi H, Hitachi K, Ohno S. Honda M, et al. J Cell Physiol. 2019 Sep;234(9):15809-15824. doi: 10.1002/jcp.28239. Epub 2019 Feb 6. J Cell Physiol. 2019. PMID: 30724341 - Transcriptional regulation of acetylcholinesterase-associated collagen ColQ: differential expression in fast and slow twitch muscle fibers is driven by distinct promoters.
Lee HH, Choi RC, Ting AK, Siow NL, Jiang JX, Massoulié J, Tsim KW. Lee HH, et al. J Biol Chem. 2004 Jun 25;279(26):27098-107. doi: 10.1074/jbc.M402596200. Epub 2004 Apr 21. J Biol Chem. 2004. PMID: 15102835 - Muscle mechanics: adaptations with exercise-training.
Fitts RH, Widrick JJ. Fitts RH, et al. Exerc Sport Sci Rev. 1996;24:427-73. Exerc Sport Sci Rev. 1996. PMID: 8744258 Review. - Gene expression in muscle in response to exercise.
Goldspink G. Goldspink G. J Muscle Res Cell Motil. 2003;24(2-3):121-6. doi: 10.1023/a:1026041228041. J Muscle Res Cell Motil. 2003. PMID: 14609023 Review.
Cited by
- Biological and therapeutic insights from animal modeling of fusion-driven pediatric soft tissue sarcomas.
Kucinski JP, Calderon D, Kendall GC. Kucinski JP, et al. Dis Model Mech. 2024 Jun 1;17(6):dmm050704. doi: 10.1242/dmm.050704. Epub 2024 Jun 25. Dis Model Mech. 2024. PMID: 38916046 Free PMC article. Review. - Cardiac biopsies reveal differences in transcriptomics between left and right ventricle in patients with or without diagnostic signs of heart failure.
Frisk C, Das S, Eriksson MJ, Walentinsson A, Corbascio M, Hage C, Kumar C, Ekström M, Maret E, Persson H, Linde C, Persson B. Frisk C, et al. Sci Rep. 2024 Mar 9;14(1):5811. doi: 10.1038/s41598-024-56025-1. Sci Rep. 2024. PMID: 38461325 Free PMC article. - DNM2 levels normalization improves muscle phenotypes of a novel mouse model for moderate centronuclear myopathy.
de Carvalho Neves J, Moschovaki-Filippidou F, Böhm J, Laporte J. de Carvalho Neves J, et al. Mol Ther Nucleic Acids. 2023 Jul 17;33:321-334. doi: 10.1016/j.omtn.2023.07.003. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37547294 Free PMC article. - VGLL2-NCOA2 leverages developmental programs for pediatric sarcomagenesis.
Watson S, LaVigne CA, Xu L, Surdez D, Cyrta J, Calderon D, Cannon MV, Kent MR, Silvius KM, Kucinski JP, Harrison EN, Murchison W, Rakheja D, Tirode F, Delattre O, Amatruda JF, Kendall GC. Watson S, et al. Cell Rep. 2023 Jan 31;42(1):112013. doi: 10.1016/j.celrep.2023.112013. Epub 2023 Jan 18. Cell Rep. 2023. PMID: 36656711 Free PMC article. - Deep learning-derived cardiovascular age shares a genetic basis with other cardiac phenotypes.
Libiseller-Egger J, Phelan JE, Attia ZI, Benavente ED, Campino S, Friedman PA, Lopez-Jimenez F, Leon DA, Clark TG. Libiseller-Egger J, et al. Sci Rep. 2022 Dec 31;12(1):22625. doi: 10.1038/s41598-022-27254-z. Sci Rep. 2022. PMID: 36587059 Free PMC article.
References
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases