Transcription Factors in Eosinophil Development and As Therapeutic Targets - PubMed (original) (raw)
Review
Transcription Factors in Eosinophil Development and As Therapeutic Targets
Patricia C Fulkerson. Front Med (Lausanne). 2017.
Abstract
Dynamic gene expression is a major regulatory mechanism that directs hematopoietic cell fate and differentiation, including eosinophil lineage commitment and eosinophil differentiation. Though GATA-1 is well established as a critical transcription factor (TF) for eosinophil development, delineating the transcriptional networks that regulate eosinophil development at homeostasis and in inflammatory states is not complete. Yet, recent advances in molecular experimental tools using purified eosinophil developmental stages have led to identifying new regulators of gene expression during eosinophil development. Herein, recent studies that have provided new insight into the mechanisms of gene regulation during eosinophil lineage commitment and eosinophil differentiation are reviewed. A model is described wherein distinct classes of TFs work together via collaborative and hierarchical interactions to direct eosinophil development. In addition, the therapeutic potential for targeting TFs to regulate eosinophil production is discussed. Understanding how specific signals direct distinct patterns of gene expression required for the specialized functions of eosinophils will likely lead to new targets for therapeutic intervention.
Keywords: eosinophil development; eosinophil lineage commitment; eosinophilopoiesis; hematopoiesis; transcriptional regulation.
Figures
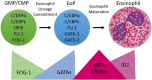
Figure 1
Transcription Factor (TF) expression during eosinophil development. Eosinophils differentiate in the bone marrow from an eosinophil lineage-committed progenitor (EoP) that is derived from the granulocyte/macrophage progenitor (GMP) in mice and the common myeloid progenitor (CMP) in humans. For eosinophil lineage commitment to occur, the myeloid progenitor (GMP or CMP) must express C/EBPα, C/EBPε, interferon regulatory factor 8 (IRF8), and PU.1. Expression of friend of GATA-1 (FOG-1) declines, allowing for increasing expression and activity of GATA TFs, which is necessary for EoP production. Following lineage commitment, eosinophil granule protein gene expression is markedly increased with the collaborative interaction between C/EBPε, PU.1, and GATA-1. To assist with the elevated granule protein synthesis in the EoP and eosinophil precursors, XBP1 expression is increased and promotes survival during the demanding maturation process. Expression of activator isoforms of C/EBPε peaks during eosinophil maturation and then declines during the final stages. Expression of ID2 increases during eosinophil maturation and enhances the rate of maturation.
Similar articles
- Transcription factor and cytokine regulation of eosinophil lineage commitment.
Mack EA, Pear WS. Mack EA, et al. Curr Opin Hematol. 2020 Jan;27(1):27-33. doi: 10.1097/MOH.0000000000000552. Curr Opin Hematol. 2020. PMID: 31688456 Free PMC article. Review. - Chromatin Preparation from Murine Eosinophils for Genome-Wide Analyses.
Bouffi C, Barski A, Fulkerson PC. Bouffi C, et al. Methods Mol Biol. 2018;1799:265-274. doi: 10.1007/978-1-4939-7896-0_20. Methods Mol Biol. 2018. PMID: 29956158 Free PMC article. - Eosinophil Development, Disease Involvement, and Therapeutic Suppression.
Fulkerson PC, Rothenberg ME. Fulkerson PC, et al. Adv Immunol. 2018;138:1-34. doi: 10.1016/bs.ai.2018.03.001. Epub 2018 Apr 22. Adv Immunol. 2018. PMID: 29731004 Review. - Cebp1 and Cebpβ transcriptional axis controls eosinophilopoiesis in zebrafish.
Li G, Sun Y, Kwok I, Yang L, Wen W, Huang P, Wu M, Li J, Huang Z, Liu Z, He S, Peng W, Bei JX, Ginhoux F, Ng LG, Zhang Y. Li G, et al. Nat Commun. 2024 Jan 27;15(1):811. doi: 10.1038/s41467-024-45029-0. Nat Commun. 2024. PMID: 38280871 Free PMC article. - Distinct C/EBP functions are required for eosinophil lineage commitment and maturation.
Nerlov C, McNagny KM, Döderlein G, Kowenz-Leutz E, Graf T. Nerlov C, et al. Genes Dev. 1998 Aug 1;12(15):2413-23. doi: 10.1101/gad.12.15.2413. Genes Dev. 1998. PMID: 9694805 Free PMC article.
Cited by
- Inflammation in Prostate Cancer: Exploring the Promising Role of Phenolic Compounds as an Innovative Therapeutic Approach.
Fernandes R, Costa C, Fernandes R, Barros AN. Fernandes R, et al. Biomedicines. 2023 Nov 24;11(12):3140. doi: 10.3390/biomedicines11123140. Biomedicines. 2023. PMID: 38137361 Free PMC article. Review. - Role of Innate Immunity in Allergic Contact Dermatitis: An Update.
Yamaguchi HL, Yamaguchi Y, Peeva E. Yamaguchi HL, et al. Int J Mol Sci. 2023 Aug 19;24(16):12975. doi: 10.3390/ijms241612975. Int J Mol Sci. 2023. PMID: 37629154 Free PMC article. Review. - Emerging functions of tissue-resident eosinophils.
Gurtner A, Crepaz D, Arnold IC. Gurtner A, et al. J Exp Med. 2023 Jul 3;220(7):e20221435. doi: 10.1084/jem.20221435. Epub 2023 Jun 16. J Exp Med. 2023. PMID: 37326974 Free PMC article. Review. - Management of eosinophil-associated inflammatory diseases: the importance of a multidisciplinary approach.
Quirce S, Cosío BG, España A, Blanco R, Mullol J, Santander C, Del Pozo V. Quirce S, et al. Front Immunol. 2023 May 17;14:1192284. doi: 10.3389/fimmu.2023.1192284. eCollection 2023. Front Immunol. 2023. PMID: 37266434 Free PMC article. - Potent CCR3 Receptor Antagonist, SB328437, Suppresses Colonic Eosinophil Chemotaxis and Inflammation in the Winnie Murine Model of Spontaneous Chronic Colitis.
Filippone RT, Dargahi N, Eri R, Uranga JA, Bornstein JC, Apostolopoulos V, Nurgali K. Filippone RT, et al. Int J Mol Sci. 2022 Jul 14;23(14):7780. doi: 10.3390/ijms23147780. Int J Mol Sci. 2022. PMID: 35887133 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous