Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins - PubMed (original) (raw)
Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins
Andreas Zanzoni et al. Microbiome. 2017.
Abstract
Background: Fusobacterium nucleatum is a gram-negative anaerobic species residing in the oral cavity and implicated in several inflammatory processes in the human body. Although F. nucleatum abundance is increased in inflammatory bowel disease subjects and is prevalent in colorectal cancer patients, the causal role of the bacterium in gastrointestinal disorders and the mechanistic details of host cell functions subversion are not fully understood.
Results: We devised a computational strategy to identify putative secreted F. nucleatum proteins (FusoSecretome) and to infer their interactions with human proteins based on the presence of host molecular mimicry elements. FusoSecretome proteins share similar features with known bacterial virulence factors thereby highlighting their pathogenic potential. We show that they interact with human proteins that participate in infection-related cellular processes and localize in established cellular districts of the host-pathogen interface. Our network-based analysis identified 31 functional modules in the human interactome preferentially targeted by 138 FusoSecretome proteins, among which we selected 26 as main candidate virulence proteins, representing both putative and known virulence proteins. Finally, six of the preferentially targeted functional modules are implicated in the onset and progression of inflammatory bowel diseases and colorectal cancer.
Conclusions: Overall, our computational analysis identified candidate virulence proteins potentially involved in the F. nucleatum-human cross-talk in the context of gastrointestinal diseases.
Keywords: Bioinformatics; Colorectal cancer; Fusobacterium nucleatum; Inflammatory bowel diseases; Interaction network; Molecular mimicry; Secretome; Short linear motifs; Virulence proteins.
Conflict of interest statement
Ethics approval and consent to participate
This study is based on publicly available datasets only. Thus, no ethical approval is needed/applicable nor is consent from any participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
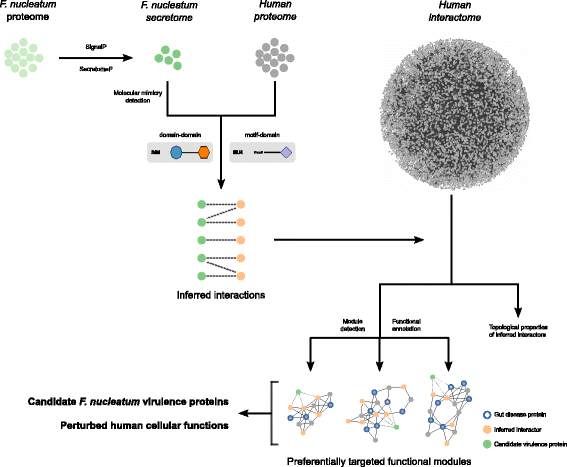
Fig. 1
Flow strategy of our computational approach
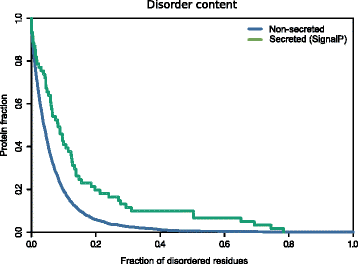
Fig. 2
Disorder propensity of the _Fuso_Secretome**.** SignalP-secreted proteins show a significantly higher fraction of disordered residues compared to non-secreted proteins (P value = 1.9 × 10−4, Kolgomorov–Smirnov test, two-sided)
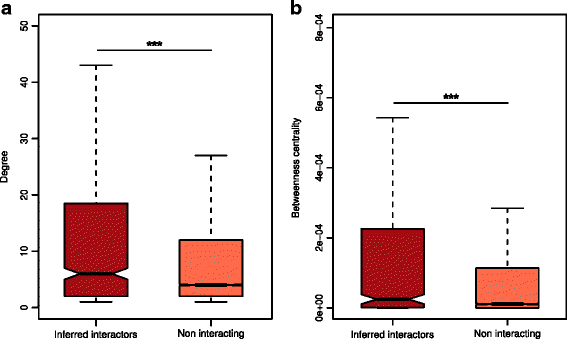
Fig. 3
Topological properties of inferred human interactors in the human interactome. a Inferred human interactors have more interaction partners and b higher values of betweenness centrality compared to non-interacting proteins in the human interactome
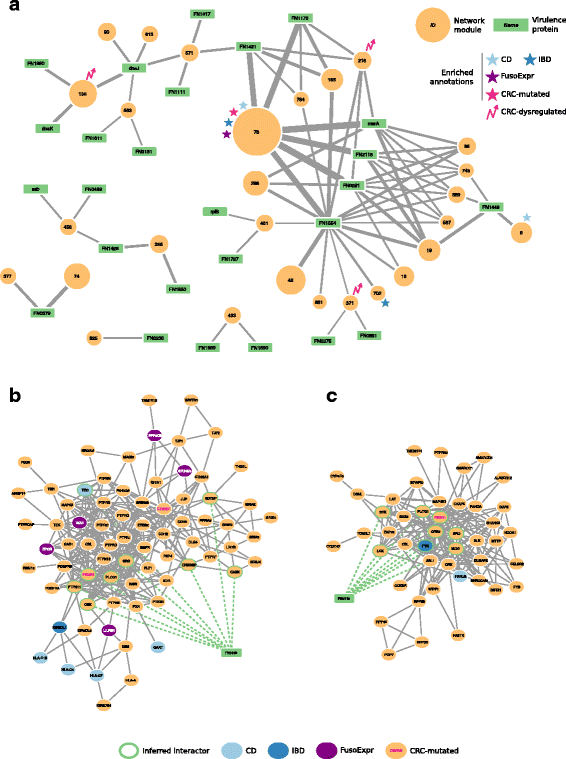
Fig. 4
Interaction network between _Fuso_Secretome candidate virulence proteins and preferentially targeted modules. a Candidate virulence proteins are depicted as green rectangular nodes labeled with respective gene symbol, whereas network modules as orange circles, whose size is proportional to the number of proteins belonging to each module and are labeled with the corresponding identifier. Edge width is proportional to the number of inferred interactions of a virulence protein with a given module. Network modules enriched in gut-related disease gene sets are labeled with symbols of different colors (i.e., light blue star: Crohn’s disease, CD; dark blue star: Inflammatory bowel disease, IBD; violet star: genes whose expression correlates with F. nucleatum abundance in colorectal cancer patients, FusoExpr; rose star: genes mutated in colorectal cancer, CRC-mutated; rose zig-zag arrow: dysregulated expression during colorectal cancer progression, CRC-dysregulated). b The protein Fap2 (FN1449) interacts with 9 proteins (nodes with a green border) of Module 9 and c the MORN2 domain containing protein (FN2118) interacts with 8 proteins in Module 89
Fig. 5
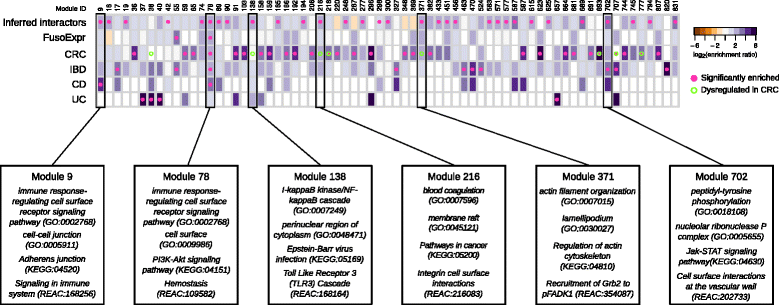
Enrichment of _Fuso_Secretome inferred human interactors and gut disease related proteins in network modules. Each column of the heatmap represents a module. The color of the cells corresponds to the log-transformed enrichment ratio. Pink circles indicate enriched sets. Modules showing a significant dysregulation in CRC progression are highlighted by an empty circle with green border. For the six modules showing an enrichment in inferred interactions and at least in one of gut disease related proteins, the most representative functions are reported. FusoExpr: genes whose expression correlates with F. nucleatum abundance in CRC patients; CRC: genes mutated in colorectal cancer samples; IBD: genes associated to inflammatory bowel disease; CD and UC: genes specifically associated to Crohn’s disease and ulcerative colitis respectively
Similar articles
- Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article. - Proteomic characterization of outer membrane vesicles from gut mucosa-derived fusobacterium nucleatum.
Liu J, Hsieh CL, Gelincik O, Devolder B, Sei S, Zhang S, Lipkin SM, Chang YF. Liu J, et al. J Proteomics. 2019 Mar 20;195:125-137. doi: 10.1016/j.jprot.2018.12.029. Epub 2019 Jan 8. J Proteomics. 2019. PMID: 30634002 - Fusobacterium nucleatum: an emerging gut pathogen?
Allen-Vercoe E, Strauss J, Chadee K. Allen-Vercoe E, et al. Gut Microbes. 2011 Sep 1;2(5):294-8. doi: 10.4161/gmic.2.5.18603. Epub 2011 Sep 1. Gut Microbes. 2011. PMID: 22067936 Review. - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review. - Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions.
Kumar A, Thotakura PL, Tiwary BK, Krishna R. Kumar A, et al. BMC Microbiol. 2016 May 12;16:84. doi: 10.1186/s12866-016-0700-0. BMC Microbiol. 2016. PMID: 27176600 Free PMC article.
Cited by
- Post-transcriptional regulatory patterns revealed by protein-RNA interactions.
Zanzoni A, Spinelli L, Ribeiro DM, Tartaglia GG, Brun C. Zanzoni A, et al. Sci Rep. 2019 Mar 13;9(1):4302. doi: 10.1038/s41598-019-40939-2. Sci Rep. 2019. PMID: 30867517 Free PMC article. - Outer Membrane Vesicles From Fusobacterium nucleatum Switch M0-Like Macrophages Toward the M1 Phenotype to Destroy Periodontal Tissues in Mice.
Chen G, Sun Q, Cai Q, Zhou H. Chen G, et al. Front Microbiol. 2022 Mar 21;13:815638. doi: 10.3389/fmicb.2022.815638. eCollection 2022. Front Microbiol. 2022. PMID: 35391731 Free PMC article. - Progress in characterizing the linkage between Fusobacterium nucleatum and gastrointestinal cancer.
Liu Y, Baba Y, Ishimoto T, Iwatsuki M, Hiyoshi Y, Miyamoto Y, Yoshida N, Wu R, Baba H. Liu Y, et al. J Gastroenterol. 2019 Jan;54(1):33-41. doi: 10.1007/s00535-018-1512-9. Epub 2018 Sep 22. J Gastroenterol. 2019. PMID: 30244399 Review. - Role of the oral microbiota in cancer evolution and progression.
Sun J, Tang Q, Yu S, Xie M, Xie Y, Chen G, Chen L. Sun J, et al. Cancer Med. 2020 Sep;9(17):6306-6321. doi: 10.1002/cam4.3206. Epub 2020 Jul 7. Cancer Med. 2020. PMID: 32638533 Free PMC article. Review. - Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources