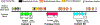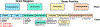Identification of the Colicin V Bacteriocin Gene Cluster by Functional Screening of a Human Microbiome Metagenomic Library - PubMed (original) (raw)
Identification of the Colicin V Bacteriocin Gene Cluster by Functional Screening of a Human Microbiome Metagenomic Library
Louis J Cohen et al. ACS Infect Dis. 2018.
Abstract
The forces that shape human microbial ecology are complex. It is likely that human microbiota, similarly to other microbiomes, use antibiotics as one way to establish an ecological niche. In this study, we use functional metagenomics to identify human microbial gene clusters that encode for antibiotic functions. Screening of a metagenomic library prepared from a healthy patient stool sample led to the identification of a family of clones with inserts that are 99% identical to a region of a virulence plasmid found in avian pathogenic Escherichia coli. Characterization of the metagenomic DNA sequence identified a colicin V biosynthetic cluster as being responsible for the observed antibiotic effect of the metagenomic clone against E. coli. This study presents a scalable method to recover antibiotic gene clusters from humans using functional metagenomics and highlights a strategy to study bacteriocins in the human microbiome which can provide a resource for therapeutic discovery.
Keywords: antibiotic; bacteriocin; eDNA; metagenome; microbiome.
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures
Figure 1.
Functional metagenomic screen for antibiotics. (A) Samples of metagenomic DNA can be isolated from any part of the human microbiome and cloned into a cosmid DNA library (B). Cosmids containing metagenomic DNA are introduced into heterologous bacterial hosts, where foreign metagenomic DNA is expressed. (C) Metagenomic clones are plated on agar and allowed to produce potential antibiotics encoded on the piece of metagenomic DNA. Indicator strains are overlaid on top of the metagenomic clones, and active metagenomic clones are identified by inhibition of growth of the indicator strain, producing a zone of clearing. Pictured is an active metagenomic clone inhibiting the growth of an E. coli indicator strain.
Figure 2.
Active metagenomic clone DNA sequence. The metagenomic DNA insert contains three gene clusters with predicted virulence functions including colicin V, colicin E9, and a protease toxin. The flanking regions of the DNA contain mobile element genes. Transposon insertion sites in the genes of the colicin V cluster are pictured.
Figure 3.
Spectrum of activity. E. coli expressing the colicin V gene cluster was plated on LYH-BHI agar media and overlaid with indicator strains of 20 human commensal bacteria from the four common phyla present in the human microbiome. Inhibition of the indicator strain was only observed for E. coli strains K12 and MS145–7, a strain of adherent invasive E. coli (AIEC), both indicated in red. There was no inhibition of any strains by E. coli carrying the empty vector.
Similar articles
- A metagenomic strategy for harnessing the chemical repertoire of the human microbiome.
Sugimoto Y, Camacho FR, Wang S, Chankhamjon P, Odabas A, Biswas A, Jeffrey PD, Donia MS. Sugimoto Y, et al. Science. 2019 Dec 13;366(6471):eaax9176. doi: 10.1126/science.aax9176. Epub 2019 Oct 3. Science. 2019. PMID: 31582523 - Identification of biosynthetic gene clusters from metagenomic libraries using PPTase complementation in a Streptomyces host.
Bitok JK, Lemetre C, Ternei MA, Brady SF. Bitok JK, et al. FEMS Microbiol Lett. 2017 Sep 1;364(16):fnx155. doi: 10.1093/femsle/fnx155. FEMS Microbiol Lett. 2017. PMID: 28817927 Free PMC article. - A rapid and efficient strategy to identify and recover biosynthetic gene clusters from soil metagenomes.
Negri T, Mantri S, Angelov A, Peter S, Muth G, Eustáquio AS, Ziemert N. Negri T, et al. Appl Microbiol Biotechnol. 2022 Apr;106(8):3293-3306. doi: 10.1007/s00253-022-11917-y. Epub 2022 Apr 18. Appl Microbiol Biotechnol. 2022. PMID: 35435454 Free PMC article. - Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities.
Suenaga H. Suenaga H. Environ Microbiol. 2012 Jan;14(1):13-22. doi: 10.1111/j.1462-2920.2011.02438.x. Epub 2011 Mar 1. Environ Microbiol. 2012. PMID: 21366818 Review. - Metagenomic approaches: effective tools for monitoring the structure and functionality of microbiomes in anaerobic digestion systems.
Carabeo-Pérez A, Guerra-Rivera G, Ramos-Leal M, Jiménez-Hernández J. Carabeo-Pérez A, et al. Appl Microbiol Biotechnol. 2019 Dec;103(23-24):9379-9390. doi: 10.1007/s00253-019-10052-5. Epub 2019 Nov 9. Appl Microbiol Biotechnol. 2019. PMID: 31420693 Review.
Cited by
- Biosynthesis of the sactipeptide Ruminococcin C by the human microbiome: Mechanistic insights into thioether bond formation by radical SAM enzymes.
Balty C, Guillot A, Fradale L, Brewee C, Lefranc B, Herrero C, Sandström C, Leprince J, Berteau O, Benjdia A. Balty C, et al. J Biol Chem. 2020 Dec 4;295(49):16665-16677. doi: 10.1074/jbc.RA120.015371. Epub 2020 Sep 24. J Biol Chem. 2020. PMID: 32972973 Free PMC article. - Success of microbial genes based transgenic crops: Bt and beyond Bt.
Tilgam J, Kumar K, Jayaswal D, Choudhury S, Kumar A, Jayaswall K, Saxena AK. Tilgam J, et al. Mol Biol Rep. 2021 Dec;48(12):8111-8122. doi: 10.1007/s11033-021-06760-9. Epub 2021 Oct 30. Mol Biol Rep. 2021. PMID: 34716867 Review. - Approaches for characterizing and tracking hospital-associated multidrug-resistant bacteria.
Blake KS, Choi J, Dantas G. Blake KS, et al. Cell Mol Life Sci. 2021 Mar;78(6):2585-2606. doi: 10.1007/s00018-020-03717-2. Epub 2021 Feb 13. Cell Mol Life Sci. 2021. PMID: 33582841 Free PMC article. Review. - Complete Genome Sequence of Acinetobacter radioresistens Strain LH6, a Multidrug-Resistant Bacteriophage-Propagating Strain.
Crippen CS, Huynh S, Miller WG, Parker CT, Szymanski CM. Crippen CS, et al. Microbiol Resour Announc. 2018 Aug 9;7(5):e00929-18. doi: 10.1128/MRA.00929-18. eCollection 2018 Aug. Microbiol Resour Announc. 2018. PMID: 30533885 Free PMC article. - Radical SAM Enzymes and Ribosomally-Synthesized and Post-translationally Modified Peptides: A Growing Importance in the Microbiomes.
Benjdia A, Berteau O. Benjdia A, et al. Front Chem. 2021 Jul 19;9:678068. doi: 10.3389/fchem.2021.678068. eCollection 2021. Front Chem. 2021. PMID: 34350157 Free PMC article. Review.
References
- Sommer F, and Backhed F (2013) The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol 11, 227–238. - PubMed
- Stappenbeck TS, and Virgin HW (2016) Accounting for reciprocal host-microbiome interactions in experimental science. Nature 534, 191–199. - PubMed
- Riley MA, and Wertz JE (2002) Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol 56, 117–137. - PubMed
- Cordero OX, et al. (2012) Ecological Populations of Bacteria Act as Socially Cohesive Units of Antibiotic Production and Resistance. Science 337, 1228–1231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources