Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency - PubMed (original) (raw)
Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency
Bin Li et al. Nat Biomed Eng. 2017 May.
Abstract
Cpf1, a type-V CRISPR-Cas effector endonuclease, exhibits gene-editing activity in human cells through a single RNA-guided approach. Here, we report the design and assessment of an array of 42 types of engineered Acidaminococcus sp. Cpf1 (AsCpf1) CRISPR RNAs (crRNAs) and 5 types of AsCpf1 mRNAs, and show that the top-performing modified crRNA (cr3'5F, containing five 2'-fluoro ribose at the 3' termini) and AsCpf1 mRNA (full ψ-modification) improved gene-cutting efficiency by, respectively, 127% and 177%, with respect to unmodified crRNA and plasmid-encoding AsCpf1. We also show that the combination of cr3'5F and ψ-modified AsCpf1 or Lachnospiraceae bacterium Cpf1 (LbCpf1) mRNAs augmented gene-cutting efficiency by over 300% with respect to the same control, and discovered that 11 out of 16 crRNAs from Cpf1 orthologs enabled genome editing in the presence of AsCpf1. Engineered CRISPR-Cpf1 systems should facilitate a broad range of genome editing applications.
Conflict of interest statement
Competing interests The authors declare no competing financial interests.
Figures
Figure 1. Design of chemically modified and structurally engineered crRNAs
a, Schematic of the AsCpf1-crRNA-target DNA complex. crRNA is composed of a 5′-handle (in a pseudoknot structure) and a guide segment (consists of a seed region and 3′-termini). The dotted line denotes the cleavage sites. b, Panel of the chemically modified crRNAs tested in this study. Unmodified nucleotides are shown in black. Full-length PS modifications are in pink. 2′-_O_-methyl modifications are highlighted in blue. 2′-F modifications are in red. 2′-_O_-methyl combined with PS modifications are in light blue. 2′-F combined with PS modifications are in green. c, Stem engineering of crRNAs. The length of the stem duplex was altered by deletion or insertion (in light orange) of nucleotides. For crSplit, the pseudoknot-like structure was formed by hybridization of two arms of the direct repeat.
Figure 2. Gene-cutting efficiency of chemically modified and stem-engineered crRNAs
a, Gene-cutting efficiency of chemically modified crRNAs. Unlock nucleotides are shown in purple. Locked nucleotides are shown in orange. Other colors in sequence are the same as that in Fig. 1b. b, Gene-cutting efficiency of stem-engineered crRNAs. “T” in the left panel denotes the split site. The dash denotes deleted nucleotide. The lowercase letter denotes inserted nucleotides. Gene-cutting efficiencies in a and b were determined by the T7E1 cleavage assay, and normalized to that of the treatment with crWT and plasmid-encoding AsCpf1. N.D., Not detectable. Data were expressed as the mean ± s.d. from three biological replicates (**, P < 0.01, cr3′5F versus to crWT; t test, double-tailed).
Figure 3. Gene-cutting efficiency of chemically modified AsCpf1 mRNAs and loop-engineered crRNAs
a, Schematic diagram depicting chemical modifications applied to CRISPR-Cpf1 mRNAs. Modified Cpf1 mRNAs were generated by fully substituting natural uridines (blue box) of unmodified mRNA (WT mRNA) with pseudouridine (ψ), N-methylpseudouridine (me1ψ), or 5-methoxyuridine (5moU). b, Gene-cutting efficiency of chemically modified AsCpf1 mRNAs. For S1228A & ψ modified AsCpf1 mRNA, the nucleotides that encode serine1228 (S1228) were substituted with that of alanine (A), and uridines were fully substituted with pseudouridine (ψ). Indel percentage was determined by the T7E1 cleavage assay, and normalized to that of the treatment with crWT and plasmid-encoding AsCpf1. N.D., Not detectable. (***, P < 0.001, ψ-modified AsCpf1 mRNA versus to AsCpf1 plasmid; t test, double-tailed). c, Loop engineering of crRNAs. Nucleotides of the loop were altered according to crRNAs from other 15 Cpf1-family orthologs. crRNAs were termed by combining the initials of the Cpf1 protein followed by crRNA; crRNAs of PbCpf1, PeCpf1 and LiCpf1 share the same loop, UUUU; crRNAs of Lb2Cpf1, PcCpf1 and PmCpf1 share the same loop, UAUU. Differences in nucleotides between AscrRNA and other crRNAs in the loop are highlighted in red. d, Gene-cutting efficiency of loop-engineered crRNAs in the presence of ψ-modified AsCpf1 mRNA. Red nucleotides denote sequence differences in the loop between AscrRNA and other crRNAs. Gene-cutting efficiency was determined by the T7E1 cleavage assay, and normalized to that of the treatment with AscrRNA and ψ-modified AsCpf1 mRNA. N.D., Not detectable. Data were expressed as the mean ± s.d. from three biological replicates (*, P < 0.05, Lb2crRNA versus to AscrRNA; t test, double-tailed).
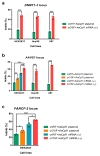
Figure 4. Maximizing genome editing efficiency through the combination of chemically modified crRNA and AsCpf1 mRNA
a, AsCpf1-mediated gene-cutting efficiency for the human DNMT1-3 in HEK293T, Hep3B and U87 cells. b, AsCpf1-mediated gene-cutting efficiency for the human AAVS1 in HEK293T, Hep3B and U87 cells. c, AsCpf1-mediated gene-cutting efficiency for the human FANCF-2 in HEK293T cells. Indel percentage at each locus was determined using the T7E1 assay, and expressed as the mean ± s.d. from three biological replicates (*, P < 0.05; **, P < 0.01; ***, P < 0.001; t test, double-tailed). Gel images are in Supplementary Fig. 3.
Figure 5. Engineered LbCpf1 crRNAs and mRNAs, and crRNA applicability
a, LbCpf1-mediated gene-cutting efficiency for the human DNMT1-3 in HEK293T, Hep3B and U87 cells. Indel percentage was determined using the T7E1 assay, and expressed as the mean ± s.d. from three biological replicates (**, P < 0.01; t test, double-tailed). Gel images are in Supplementary Fig. 4. b, Gene-cutting efficiency of loop-engineered crRNAs in the presence of ψ-modified LbCpf1 mRNA. Red nucleotides denote sequence difference in the loop between LbcrRNA and other crRNAs. Gene-cutting efficiency was determined by the T7E1 cleavage assay, normalized to that of the treatment with LbcrRNA and ψ-modified LbCpf1 mRNA, and expressed as the mean ± s.d. from three biological replicates. Gel images are in Supplementary Fig. 4.
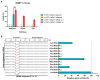
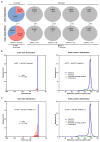
Figure 6. Targeted-deep-sequencing analysis of on-target and off-target gene cutting for chemically modified crRNA and AsCpf1 mRNA
a, Indel percentage at on-target and predicted top four off-target sites analyzed by deep-sequencing data. Indel percentage was plotted as the mean ± s.d. from three biological replicates (***, P < 0.001; N.S.; Not significant; t test, double-tailed). b, Plot of representative indel-size distribution and position distribution of all reads for crWT + AsCpf1 plasmid. c, Representative plots of indel-size distribution and position distribution of all reads for cr3′5F+ ψ-modified AsCpf1 mRNA. Targeted-deep-sequencing analysis for biological replicates 2 and 3 are in Supplementary Fig. 7 and 8.
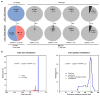
Figure 7. Targeted-deep-sequencing analysis of on-target and off-target gene cutting for chemically modified Lbcr3
′5F and LbCpf1 mRNA. a, Indel percentage at on-target and predicted top four off-target sites analyzed by deep-sequencing data. Indel percentage was plotted as the mean ± s.d. from three biological replicates (***, P < 0.001; N.S.; Not significant; t test, double-tailed). b, Representative plots of indel-size distribution and position distribution of all reads for Lbcr3′5F + ψ-modified LbCpf1 mRNA. Targeted-deep-sequencing analysis for biological replicates 2 and 3 are in Supplementary Fig. 7 and 8.
Similar articles
- CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae.
Verwaal R, Buiting-Wiessenhaan N, Dalhuijsen S, Roubos JA. Verwaal R, et al. Yeast. 2018 Feb;35(2):201-211. doi: 10.1002/yea.3278. Epub 2017 Nov 12. Yeast. 2018. PMID: 28886218 Free PMC article. - Efficient genome editing in wheat using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases.
Kim D, Hager M, Brant E, Budak H. Kim D, et al. Funct Integr Genomics. 2021 Jul;21(3-4):355-366. doi: 10.1007/s10142-021-00782-z. Epub 2021 Mar 12. Funct Integr Genomics. 2021. PMID: 33710467 - Efficient multiplex CRISPR/Cpf1 (Cas12a) genome editing system in Aspergillus aculeatus TBRC 277.
Abdulrachman D, Champreda V, Eurwilaichitr L, Chantasingh D, Pootanakit K. Abdulrachman D, et al. J Biotechnol. 2022 Aug 20;355:53-64. doi: 10.1016/j.jbiotec.2022.06.011. Epub 2022 Jul 2. J Biotechnol. 2022. PMID: 35788357 - CRISPR-Cpf1-mediated genome editing and gene regulation in human cells.
Li T, Zhu L, Xiao B, Gong Z, Liao Q, Guo J. Li T, et al. Biotechnol Adv. 2019 Jan-Feb;37(1):21-27. doi: 10.1016/j.biotechadv.2018.10.013. Epub 2018 Nov 3. Biotechnol Adv. 2019. PMID: 30399413 Review. - The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants.
Alok A, Sandhya D, Jogam P, Rodrigues V, Bhati KK, Sharma H, Kumar J. Alok A, et al. Front Plant Sci. 2020 Mar 31;11:264. doi: 10.3389/fpls.2020.00264. eCollection 2020. Front Plant Sci. 2020. PMID: 32296449 Free PMC article. Review.
Cited by
- Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system.
Wang L, Han H. Wang L, et al. Heliyon. 2024 Sep 27;10(19):e38588. doi: 10.1016/j.heliyon.2024.e38588. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397905 Free PMC article. Review. - Cas12a and MAD7, genome editing tools for breeding.
Hozumi S, Chen YC, Takemoto T, Sawatsubashi S. Hozumi S, et al. Breed Sci. 2024 Mar;74(1):22-31. doi: 10.1270/jsbbs.23049. Epub 2024 Feb 22. Breed Sci. 2024. PMID: 39246434 Free PMC article. - Harnessing noncanonical crRNA for highly efficient genome editing.
Xun G, Zhu Z, Singh N, Lu J, Jain PK, Zhao H. Xun G, et al. Nat Commun. 2024 May 7;15(1):3823. doi: 10.1038/s41467-024-48012-x. Nat Commun. 2024. PMID: 38714643 Free PMC article. - Self-delivering, chemically modified CRISPR RNAs for AAV co-delivery and genome editing in vivo.
Zhang H, Kelly K, Lee J, Echeverria D, Cooper D, Panwala R, Amrani N, Chen Z, Gaston N, Wagh A, Newby GA, Xie J, Liu DR, Gao G, Wolfe SA, Khvorova A, Watts JK, Sontheimer EJ. Zhang H, et al. Nucleic Acids Res. 2024 Jan 25;52(2):977-997. doi: 10.1093/nar/gkad1125. Nucleic Acids Res. 2024. PMID: 38033325 Free PMC article. - CRISPR/Cas12a-based approaches for efficient and accurate detection of Phytophthora ramorum.
Guo Y, Xia H, Dai T, Liu T, Shamoun SF, CuiPing W. Guo Y, et al. Front Cell Infect Microbiol. 2023 Jun 27;13:1218105. doi: 10.3389/fcimb.2023.1218105. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37441240 Free PMC article.
References
- Bosley KS, et al. CRISPR germline engineering--the community speaks. Nat Biotechnol. 2015;33:478–486. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources