Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells - PubMed (original) (raw)
. 2017 Dec;18(12):776-790.
doi: 10.1111/tra.12526. Epub 2017 Oct 16.
Affiliations
- PMID: 28869701
- PMCID: PMC5677552
- DOI: 10.1111/tra.12526
Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells
Rebecca L Adams et al. Traffic. 2017 Dec.
Erratum in
- Correction to: Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells.
Adams RL, Mason AC, Glass L, Aditi, Wente SR. Adams RL, et al. Traffic. 2018 Aug;19(8):650. doi: 10.1111/tra.12585. Traffic. 2018. PMID: 29999599 No abstract available.
Abstract
The mRNA lifecycle is driven through spatiotemporal changes in the protein composition of mRNA particles (mRNPs) that are triggered by RNA-dependent DEAD-box protein (Dbp) ATPases. As mRNPs exit the nuclear pore complex (NPC) in Saccharomyces cerevisiae, this remodeling occurs through activation of Dbp5 by inositol hexakisphosphate (IP6 )-bound Gle1. At the NPC, Gle1 also binds Nup42, but Nup42's molecular function is unclear. Here we employ the power of structure-function analysis in S. cerevisiae and human (h) cells, and find that the high-affinity Nup42-Gle1 interaction is integral to Dbp5 (hDDX19B) activation and efficient mRNA export. The Nup42 carboxy-terminal domain (CTD) binds Gle1/hGle1B at an interface distinct from the Gle1-Dbp5/hDDX19B interaction site. A nup42-CTD/gle1-CTD/Dbp5 trimeric complex forms in the presence of IP6 . Deletion of NUP42 abrogates Gle1-Dbp5 interaction, and disruption of the Nup42 or IP6 binding interfaces on Gle1/hGle1B leads to defective mRNA export in S. cerevisiae and human cells. In vitro, Nup42-CTD and IP6 stimulate Gle1/hGle1B activation of Dbp5 and DDX19B recombinant proteins in similar, nonadditive manners, demonstrating complete functional conservation between humans and S. cerevisiae. Together, a highly conserved mechanism governs spatial coordination of mRNP remodeling during export. This has implications for understanding human disease mutations that perturb the Nup42-hGle1B interaction.
Keywords: mRNA export; mRNP remodeling; Dbp5; Gle1; Nup42; inositol hexakisphosphate; nuclear pore complex.
© 2017 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
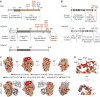
Figure 1. Gle1-Nup42 interaction domains
(A) Domain structure of Gle1, hGle1B, and hGle1A. For Gle1, K377 K378 indicates IP6-coordinating residues identified in, V513 A516 I520 indicates residues essential for Dbp5 ATPase regulation, and Q491 K494 E501 E502 indicates residues essential for interaction with Nup42 identified in this paper. For hGle1B, K526 K527 are homologous to S. cerevisiae Gle1 K377 K378, with K479 K486 as additional positively-charged residues at the IP6 interface (based on homologous residues in the Gle1 structure, PDB 3RRM), and Q640 K643 E650 D651 indicates residues essential for interaction with hNup42 identified in this paper. Symbols denote changes from disease-associated mutations,: asterisks are ALS-associated mutations, filled circle indicates LCCS1 Finmajor, open circles indicate other LCCS1 or LAAHD mutations. From amino to carboxy-terminus: S70X, ALS-associated nonsense mutation; T144_E145insPFQ, LCCS1 Finmajor; R569H, LCCS1; V617M, LAAHD; 654/IVS14-2A>C, ALS (alternate exon use results in novel C-terminal extension from this residue); I694T, LAAHD; R697C, ALS. (B) Domain structure of Nup42 and hNup42. For Nup42, 408–424 indicates minimal Gle1 interaction domain defined in this paper. (A & B) Colored regions indicate truncations used in biochemistry experiments. (C) S. cerevisiae Gle1 contains a conserved patch of residues opposite the Dbp5 binding site. From a Clustal Omega sequence alignment between Gle1 and hGle1B, identical (red) and similar (orange) residues were mapped onto the Gle1-Dbp5 structure, PDB 3RRM). (D) Q491, K494, E501, and E502 are surface accessible residues in the conserved patch on S. cerevisiae Gle1. Residues are indicated on zoomed region boxed in C.
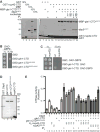
Figure 2. Functional analysis of the proposed Nup42 binding site on Gle1
(A) QKEE>AAAA disrupts the Gle1-Nup42 Y2H interaction. Indicated plasmids were transformed into the Y2H reporter strain, grown to early log phase at 23°C, and plated on the synthetic media lacking indicated amino acids for growth at 23°C. (B) gle1QKEE>AAAA ipk1Δ phenocopies the growth defect of nup42Δ ipk1Δ. Indicated strains were grown to mid-log phase, serially diluted, and plated on YPD for growth at the indicated temperatures. (C) Gle1 is localized at NPCs when interaction with Nup42 is disrupted. Indicated strains containing carboxy-terminal GFP-tagged GLE1-GFP plasmids were grown to mid-log phase in YPD at 23°C and live cells were imaged by wide-field microscopy. Scale bar = 5μm. (D) gle1QKEE>AAAA disrupts heat-shock mRNA export. Indicated strains were grown at 25°C to early-log phase, kept at 25°C or shifted to 42°C for 15 min, labeled with [35S]methionine for an additional 15 min, and lysed. Lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. The positions of Hsp proteins, induced upon heat shock, are indicated by asterisks.
Figure 3. Analysis of minimal Gle1-binding domain in Nup42
(A) nup42408–424 is required for interaction with Gle1. Indicated plasmids were transformed into the Y2H reporter strain, and struck to –Trp –Leu –His –Ade synthetic media for growth at 23°C. Growth on quadruple drop-out media is indicated as “+”. (B) nup42408–424 lies in a conserved region of the protein. Clustal Omega alignment between nup42-CTD and hnup42-CTD. Identical (black) and similar (grey) residues are indicated. The black line above the sequence indicates residues 408–424 of Nup42. (C) Affinity measurements between gle1-CTD and nup42408–424. BioLayer Interferometry was performed with a biotin-nup42408–424 peptide and recombinant gle1-CTD protein. Calculated KD and R2 correlation is indicated. (D) GFP-nup42408–424 localization at the nuclear envelope is disrupted in gle1QKEE>AAAA mutants. Strains containing the indicated plasmids were grown to mid-log phase at 23°C and live cells were imaged by wide-field microscopy. Scale bar = 5μm. (E) nup42408–424 does not rescue the growth defect of nup42Δ ipk1Δ mutants. nup42Δ ipk1Δ was transformed with the indicated GFP-tagged constructs, grown to mid-log phase, and plated on –Leu synthetic media for growth at the indicated temperatures. (F) nup42408–424 is not sufficient for heat shock mRNA export. nup42Δ mutants were transformed with the indicated GFP-tagged constructs, grown at 25°C to early-log phase in synthetic media, kept at 25°C or shifted to 42°C for 15 min, labeled with [35S]methionine for an additional 15 min, and lysed. Lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. Hsp proteins are indicated by asterisks.
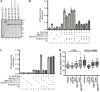
Figure 4. Nup42 coordinates Gle1-Dbp5 interaction
(A) Nup42, Gle1, and Dbp5 interact in a trimeric protein complex with IP6. Soluble binding assays were conducted with purified recombinant proteins immobilized on glutathione coupled Sepharose. Input and bound fractions were analyzed by SDS-PAGE and Coomassie staining. Note: GST-nup42-CTD is proteolytically sensitive and resolves as several bands by SDS-PAGE (lane 2), as observed in. (B) GBD-gle1-CTDQKEE>AAAA does not interact with GAD-Dbp5 via Y2H. Indicated plasmids were transformed into the Y2H reporter strain, grown to early log phase at 23°C, and plated on the synthetic media lacking indicated amino acids for growth at 23°C. (C) GBD-gle1-CTD does not interact with GAD-Dbp5 in nup42Δ mutants. Indicated plasmids were transformed into wild-type (wt) or nup42Δ Y2H reporter strains, grown to early log phase at 23°, and plated on the synthetic media lacking indicated amino acids for growth at 23°C. (D) Purified recombinant S. cerevisiae proteins used in ATPase assays. 1μg indicated proteins were resolved by SDS-PAGE and Coomassie stained. Asterisk indicates GST cleaved (not removed) from Dbp5 sample. (E) nup42-CTD enhances Gle1 stimulation of Dbp5 ATPase activity in the absence of IP6. PK/LDH-coupled ATPase assays were performed with Dbp5 (500 nM) in the presence of 1 μM A30 RNA and 2 mM ATP gle1-CTD or gle1-CTDQKEE>AAAA (250 nM), IP6 (1 μM), and varying amounts of nup42-CTD or nup42408–424 peptide (250, 500, or 750 nM) were added as indicated. Reactions were incubated at 37°C for 40 min with the A340 monitored every 40 seconds to calculate Kcat. Mean shown for n=3, and standard error is indicated by error bars.
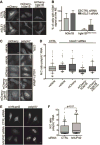
Figure 5. hGle1B interaction with hNup42 is required for mRNA export
(A–B) mCherry-hgle1BQKED>AAAA is localized to the nuclear rim in a portion of cells. (A) HeLa cells expressing exogenous siRNA-resistant mCherry-hGLE1BR and mCherry-hgle1BR-QKED>AAAA after treatment with CTRL or hGLE1 siRNA were imaged by confocal microscopy. Scale bar=10μm. (B) Quantification of cells with rim-localized mCherry signal from the experiment in A. Approximately sixty cells were quantified from three independent experiments, and the mean is indicated with standard error indicated by error bars. (C-D) mCherry-hgle1BQKED>AAAA does not rescue the mRNA export defect upon hGLE1 knockdown. (C) HeLa cells were transfected with mCherry, mCherry-hGLE1BR or mCherry-hgle1BR-QKED>AAAA plasmids following CTRL or hGLE1 knockdown. After fixation, cells were processed for in situ hybridization using 488-labeled oligo d(T) probe followed by mCherry immunofluoresence. Scale bar=10μm. (D) Quantification of nuclear:cytoplasmic mean fluorescence intensity of oligo d(T) signal. Approximately 100 cells were quantified per condition from three independent experiments with nuclear:cytoplasmic intensity ratio depicted in box and whisker plots (whiskers representing 10%–90%). Statistical analysis includes ANOVA with a post-hoc t-test. (E-F) hNUP42 depletion results in a poly(A)+ mRNA export defect. (E) HeLa cells were transfected with CTRL or hNUP42 siRNA for 72h. After fixation, cells were processed for in situ hybridization using Cy3-labeled oligo d(T) probe followed by hNup42 immunofluorescence. Scale bar=10μm. (F) Quantification of nuclear:cytoplasmic mean fluorescence intensity of oligo d(T) signal. Approximately 200 cells were quantified per condition from three independent experiments with nuclear:cytoplasmic intensity ratio depicted in box and whisker plots (whiskers representing 10%–90%). A t-test was performed to analyze statistical significance.
Figure 6. hGle1B interactions with hNup42 and IP6 enhances stimulation of DDX19B activity and are required for mRNA export
(A) Purified recombinant human proteins used in ATPase assays. 1μg indicated purified proteins were resolved by SDS-PAGE and Coomassie stained. (B) hGle1B and hNup42 stimulate DDX19B ATPase activity in absence of IP6. PK/LDH-coupled ATPase assays were performed with H6-DDX19B (500 nM, expressed in insect cells) in the presence of 1 μM A30 RNA and 2 mM ATP. hgle1B-CTD or hgle1B-CTDQKED>AAAA (250 nM), IP6 (1 μM), and varying amounts of hnup42-CTD (250, 500, or 750 nM) were added as indicated. Reactions preformed as in Figure 3D. Mean shown for n=3, and standard error is indicated by error bars. (C) hGle1B-IP6 interaction is conserved in human proteins. PK/LDH-coupled ATPase assays were performed with H6-DDX19B (500 nM) in the presence of 1 μM A30 RNA and 2 mM ATP, hgle1B-CTD, hgle1B-CTDKK>QQ, or hgle1B-CTDKKKK>QQQQ (250 nM), and hnup42-CTD (750 nM) or IP6 (1 μM) were added as indicated. Purified proteins are shown in 6A. Reactions performed as in Figure 3D. Mean shown for n=3, and standard error is indicated by error bars. (D) hGle1B-IP6 interaction is required for mRNA export. HeLa cells were transfected with vehicle or GFP-hGLE1BR, GFP-hgle1BR-KK>QQ, or GFP-hgle1BR-KKKK>QQQQ plasmids following CTRL or hGLE1 knockdown. Nuclear:cytoplasmic mean fluorescence intensity of oligo d(T) signal was quantified as in Figure 5F.
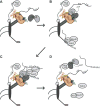
Figure 7. Schematic for Nup42 function at the NPC cytoplasmic face
(A) Dbp5 dynamically associates with the NPC. (B) The Nup42 CTD stimulates Gle1-IP6 interaction with Dbp5, with Gle1-IP6 promoting ATP loading onto Dbp5. Mature mRNPs interact with the Mex67-Mtr2 heterodimer via adaptors such as the poly(A)+ binding protein Nab2, permitting export through the NPC. (C) The FG domain of Nup42 recruits the mRNP in close proximity to Dbp5, and Dbp5-ATP binds RNA. Dbp5 might also be a constituent of the exporting mRNP. (D) ATP hydrolysis induces a conformational change in Dbp5 and the bound RNA for release of Nab2, and Mex67-Mtr2, which are recycled into the nucleus for additional rounds of mRNP export. The remodeled mRNP is released into the cytoplasm. Dbp5 interacts with the Nup159 amino terminal domain (NTD) to facilitate ADP release.
Similar articles
- Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export.
Alcázar-Román AR, Tran EJ, Guo S, Wente SR. Alcázar-Román AR, et al. Nat Cell Biol. 2006 Jul;8(7):711-6. doi: 10.1038/ncb1427. Epub 2006 Jun 18. Nat Cell Biol. 2006. PMID: 16783363 - Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export.
Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Weirich CS, et al. Nat Cell Biol. 2006 Jul;8(7):668-76. doi: 10.1038/ncb1424. Epub 2006 Jun 18. Nat Cell Biol. 2006. PMID: 16783364 - Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex.
Adams RL, Terry LJ, Wente SR. Adams RL, et al. Genetics. 2014 Aug;197(4):1213-24. doi: 10.1534/genetics.114.164012. Epub 2014 Jun 14. Genetics. 2014. PMID: 24931410 Free PMC article. - Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export.
Folkmann AW, Noble KN, Cole CN, Wente SR. Folkmann AW, et al. Nucleus. 2011 Nov-Dec;2(6):540-8. doi: 10.4161/nucl.2.6.17881. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064466 Free PMC article. Review. - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
- The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration.
Ferreira PA. Ferreira PA. Cell Mol Life Sci. 2019 Jun;76(12):2247-2273. doi: 10.1007/s00018-019-03029-0. Epub 2019 Feb 11. Cell Mol Life Sci. 2019. PMID: 30742233 Free PMC article. Review. - Snx4-assisted vacuolar targeting of transcription factors defines a new autophagy pathway for controlling ATG expression.
Hanley SE, Willis SD, Cooper KF. Hanley SE, et al. Autophagy. 2021 Nov;17(11):3547-3565. doi: 10.1080/15548627.2021.1877934. Epub 2021 Mar 8. Autophagy. 2021. PMID: 33678121 Free PMC article. - Emerging molecular functions and novel roles for the DEAD-box protein Dbp5/DDX19 in gene expression.
Arul Nambi Rajan A, Montpetit B. Arul Nambi Rajan A, et al. Cell Mol Life Sci. 2021 Mar;78(5):2019-2030. doi: 10.1007/s00018-020-03680-y. Epub 2020 Nov 17. Cell Mol Life Sci. 2021. PMID: 33205304 Free PMC article. Review. - A nuclear role for the DEAD-box protein Dbp5 in tRNA export.
Lari A, Arul Nambi Rajan A, Sandhu R, Reiter T, Montpetit R, Young BP, Loewen CJ, Montpetit B. Lari A, et al. Elife. 2019 Aug 27;8:e48410. doi: 10.7554/eLife.48410. Elife. 2019. PMID: 31453808 Free PMC article. - Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, Hoelz A. Lin DH, et al. Nat Commun. 2018 Jun 13;9(1):2319. doi: 10.1038/s41467-018-04459-3. Nat Commun. 2018. PMID: 29899397 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials