Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition - PubMed (original) (raw)
Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition
Katherine M Davis et al. Proc Natl Acad Sci U S A. 2017.
Abstract
Posttranslational modification of ribosomally synthesized peptides provides an elegant means for the production of biologically active molecules known as RiPPs (ribosomally synthesized and posttranslationally modified peptides). Although the leader sequence of the precursor peptide is often required for turnover, the exact mode of recognition by the modifying enzymes remains unclear for many members of this class of natural products. Here, we have used X-ray crystallography and computational modeling to examine the role of the leader peptide in the biosynthesis of a homolog of streptide, a recently identified peptide natural product with an intramolecular lysine-tryptophan cross-link, which is installed by the radical _S_-adenosylmethionine (SAM) enzyme, StrB. We present crystal structures of SuiB, a close ortholog of StrB, in various forms, including apo SuiB, SAM-bound SuiB, and a complex of SuiB with SAM and its peptide substrate, SuiA. Although the N-terminal domain of SuiB adopts a typical RRE (RiPP recognition element) motif, which has been implicated in precursor peptide recognition, we observe binding of the leader peptide in the catalytic barrel rather than the N-terminal domain. Computational simulations support a mechanism in which the leader peptide guides posttranslational modification by positioning the cross-linking residues of the precursor peptide within the active site. Together the results shed light onto binding of the precursor peptide and the associated conformational changes needed for the formation of the unique carbon-carbon cross-link in the streptide family of natural products.
Keywords: RRE domain; RiPP; SPASM domain; radical SAM enzyme; streptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
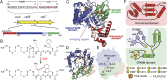
The sui gene cluster and the reaction catalyzed by SuiB. (A) The sui gene cluster is highly homologous to str. It encodes a 22-mer precursor peptide (SuiA), a tailoring radical SAM enzyme (SuiB), and a putative transporter/protease (SuiC). The sequence of SuiA is depicted with the 8-mer sequence of the mature product shown in bold. Cross-linked residues are shown in red. (B) SuiB catalyzes Lys–Trp cross-link formation in SuiA. The new bond installed is shown in red. (C) SuiB contains three [4Fe–4S] clusters and multiple functional domains. (D) The radical SAM domain (blue) forms a partial TIM barrel that is laterally closed by the auxiliary cluster-containing SPASM domain (green). The canonical catalytic [4Fe–4S] cluster-binding motif following β1 is shown in yellow. The bridging region and N-terminal domains are shown in light blue and red, respectively. (E) Placement of the three [4Fe–4S] clusters, shown in ball and stick representation (Fe, orange; S, yellow). Distances are calculated between the nearest atoms. (F) Topologies of the N-terminal domain, SPASM domain, and bridging region.
Fig. 2.
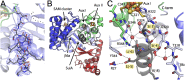
SuiA recognition in the active site is dominated by interactions of the leader sequence with the bridging region. (A) Observed electron density for the helical leader sequence displayed as a 2FO-FC composite omit map contoured at 1.0 σ. The disordered core sequence is represented by a dashed line. (B) SuiA (dark gray) binds in the catalytic barrel rather than to the putative recognition element, the N-terminal domain (red). (C) Hydrogen-bonding network of SuiA (dark gray) bound in the active site adjacent to the bridging region (light blue) and SPASM domain (green). Ordered water molecules are shown as red spheres. The LESS motif is highlighted. See SI Appendix, Fig. S5_A_ for a stereoview.
Fig. 3.
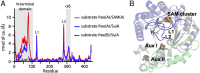
Substrate binding leads to coordinated loop movements. (A) Rmsd of the Cα atoms versus residue number upon SAM and SAM + SuiA binding, respectively. The two chains (A/B) in the asymmetric unit are denoted parenthetically. Binding only SAM leads to minimal changes (gray curve), yielding an average Cα rmsd of 0.285 ± 0.037 Å, whereas additional binding of SuiA leads to greater changes (blue/red), particularly in the loops L1 and L2. See SI Appendix, Fig. S6 for additional views. (B) Visualization of loop motions upon SuiA binding. The substrate-free enzyme is shown in gray. The RRE is omitted for clarity.
Fig. 4.
Binding of the SuiA leader sequence supports positioning of the core sequence in the active site of SuiB. (A) The arrangement of Met and SAM (green) in our SuiA-bound structure mimics previously observed SAM-cleavage products from RlmN (pink) (–31). The 5′-C of 5′-dA is marked with an asterisk. (B_–_D) The active-site cavity of SuiB is shown as a white surface. The crystallographic model of the leader peptide is shown in gray. Rosetta-based simulations yield low-energy conformations of the core sequence (blue) within our SuiA-bound crystal structure both with (B) intact SAM and (C) the methionine moiety of SAM removed. (D) Simulations also yield low-energy conformations of the cyclized core peptide (blue) within the active-site cavity. (E) Of the five titratable residues (shown as sticks) near the active site, simulations favor E319 (∼4.9 Å from the SuiA-W6 C7 position) as the catalytic base.
Similar articles
- Trapping a cross-linked lysine-tryptophan radical in the catalytic cycle of the radical SAM enzyme SuiB.
Balo AR, Caruso A, Tao L, Tantillo DJ, Seyedsayamdost MR, Britt RD. Balo AR, et al. Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2101571118. doi: 10.1073/pnas.2101571118. Proc Natl Acad Sci U S A. 2021. PMID: 34001621 Free PMC article. - Mechanistic Investigations of Lysine-Tryptophan Cross-Link Formation Catalyzed by Streptococcal Radical S-Adenosylmethionine Enzymes.
Schramma KR, Forneris CC, Caruso A, Seyedsayamdost MR. Schramma KR, et al. Biochemistry. 2018 Jan 30;57(4):461-468. doi: 10.1021/acs.biochem.7b01147. Epub 2018 Jan 23. Biochemistry. 2018. PMID: 29320164 - Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
Grove TL, Himes PM, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, Almo SC, Bowers AA. Grove TL, et al. J Am Chem Soc. 2017 Aug 30;139(34):11734-11744. doi: 10.1021/jacs.7b01283. Epub 2017 Aug 21. J Am Chem Soc. 2017. PMID: 28704043 Free PMC article. - At the confluence of ribosomally synthesized peptide modification and radical _S_-adenosylmethionine (SAM) enzymology.
Latham JA, Barr I, Klinman JP. Latham JA, et al. J Biol Chem. 2017 Oct 6;292(40):16397-16405. doi: 10.1074/jbc.R117.797399. Epub 2017 Aug 22. J Biol Chem. 2017. PMID: 28830931 Free PMC article. Review. - Structural features and substrate engagement in peptide-modifying radical SAM enzymes.
Cheek LE, Zhu W. Cheek LE, et al. Arch Biochem Biophys. 2024 Jun;756:110012. doi: 10.1016/j.abb.2024.110012. Epub 2024 Apr 23. Arch Biochem Biophys. 2024. PMID: 38663796 Review.
Cited by
- Sequence-function space of radical SAM cyclophane synthases reveal conserved active site residues that influence substrate specificity.
Phan CS, Morinaka BI. Phan CS, et al. RSC Chem Biol. 2024 Oct 23;5(12):1195-200. doi: 10.1039/d4cb00227j. Online ahead of print. RSC Chem Biol. 2024. PMID: 39464308 Free PMC article. - Biosynthesis of GMGT lipids by a radical SAM enzyme associated with anaerobic archaea and oxygen-deficient environments.
Li Y, Yu T, Feng X, Zhao B, Chen H, Yang H, Chen X, Zhang XH, Anderson HR, Burns NZ, Zeng F, Tao L, Zeng Z. Li Y, et al. Nat Commun. 2024 Jun 19;15(1):5256. doi: 10.1038/s41467-024-49650-x. Nat Commun. 2024. PMID: 38898040 Free PMC article. - Integrated sequence and -omic features reveal novel small proteome of Mycobacterium tuberculosis.
Sinha PR, Balasubramanian R, Hegde SR. Sinha PR, et al. Front Microbiol. 2024 May 15;15:1335310. doi: 10.3389/fmicb.2024.1335310. eCollection 2024. Front Microbiol. 2024. PMID: 38812687 Free PMC article. - Hydrogen-Deuterium Exchange Mass Spectrometry Identifies Local and Long-Distance Interactions within the Multicomponent Radical SAM Enzyme, PqqE.
Zhu W, Iavarone AT, Klinman JP. Zhu W, et al. ACS Cent Sci. 2024 Jan 17;10(2):251-263. doi: 10.1021/acscentsci.3c01023. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435514 Free PMC article. - Disordered regions in proteusin peptides guide post-translational modification by a flavin-dependent RiPP brominase.
Nguyen NA, Vidya FNU, Yennawar NH, Wu H, McShan AC, Agarwal V. Nguyen NA, et al. Nat Commun. 2024 Feb 10;15(1):1265. doi: 10.1038/s41467-024-45593-5. Nat Commun. 2024. PMID: 38341413 Free PMC article.
References
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 GM100008/GM/NIGMS NIH HHS/United States
- K99 GM098299/GM/NIGMS NIH HHS/United States
- P41 GM103485/GM/NIGMS NIH HHS/United States
- K99 GM100008/GM/NIGMS NIH HHS/United States
- R00 GM098299/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources