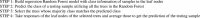Heterogeneity Aware Random Forest for Drug Sensitivity Prediction - PubMed (original) (raw)
Heterogeneity Aware Random Forest for Drug Sensitivity Prediction
Raziur Rahman et al. Sci Rep. 2017.
Abstract
Samples collected in pharmacogenomics databases typically belong to various cancer types. For designing a drug sensitivity predictive model from such a database, a natural question arises whether a model trained on diverse inter-tumor heterogeneous samples will perform similar to a predictive model that takes into consideration the heterogeneity of the samples in model training and prediction. We explore this hypothesis and observe that ensemble model predictions obtained when cancer type is known out-perform predictions when that information is withheld even when the samples sizes for the former is considerably lower than the combined sample size. To incorporate the heterogeneity idea in the commonly used ensemble based predictive model of Random Forests, we propose Heterogeneity Aware Random Forests (HARF) that assigns weights to the trees based on the category of the sample. We treat heterogeneity as a latent class allocation problem and present a covariate free class allocation approach based on the distribution of leaf nodes of the model ensemble. Applications on CCLE and GDSC databases show that HARF outperforms traditional Random Forest when the average drug responses of cancer types are different.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
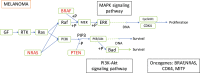
Figure 1
Melanoma (Skin) tumorigenesis pathway, collected from KEGG.
Algorithm 1
Algorithmic representation of Heterogeneity Aware Random Forest (HARF) Regression.
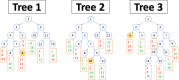
Figure 2
3 sample trees with leaf information. Boxed numbers represent the samples contained within each leaf node. Red samples belong to cancer type C A while green samples belong to cancer type C B.
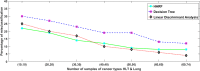
Figure 3
With an increase in the number of samples for training, the percentage of mis-classifications for HARF, Decision Tree and Linear Discriminant Analysis (LDA) all get reduced. Using drug Nilotinib of CCLE database and 2 cancer types HLT and Lung, this reduction of misclassification is shown. For small number of samples, HARF has the lowest misclassification rate. For large sample sizes, LDA gives the lowest misclassification rate, but the differences are minimal in both the cases.
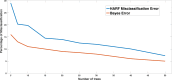
Figure 4
Changes in misclassification rate of HARF and Bayes error (Eq. 16) for different number of trees are shown. For model with few trees, misclassification rate is higher compared to model with high number of trees. As expected, HARF misclassification rate is always higher compared to minimum Bayes error, but the difference is always minimal for models with different number of trees. Drug AZD − 6244 and cancer types Skin & CNS are used for the generation of these curves.
Similar articles
- Application of transfer learning for cancer drug sensitivity prediction.
Dhruba SR, Rahman R, Matlock K, Ghosh S, Pal R. Dhruba SR, et al. BMC Bioinformatics. 2018 Dec 28;19(Suppl 17):497. doi: 10.1186/s12859-018-2465-y. BMC Bioinformatics. 2018. PMID: 30591023 Free PMC article. - Functional random forest with applications in dose-response predictions.
Rahman R, Dhruba SR, Ghosh S, Pal R. Rahman R, et al. Sci Rep. 2019 Feb 7;9(1):1628. doi: 10.1038/s41598-018-38231-w. Sci Rep. 2019. PMID: 30733524 Free PMC article. - Random forests ensemble classifier trained with data resampling strategy to improve cardiac arrhythmia diagnosis.
Ozçift A. Ozçift A. Comput Biol Med. 2011 May;41(5):265-71. doi: 10.1016/j.compbiomed.2011.03.001. Epub 2011 Mar 17. Comput Biol Med. 2011. PMID: 21419401 - Genomic approach towards personalized anticancer drug therapy.
Midorikawa Y, Tsuji S, Takayama T, Aburatani H. Midorikawa Y, et al. Pharmacogenomics. 2012 Jan;13(2):191-9. doi: 10.2217/pgs.11.157. Pharmacogenomics. 2012. PMID: 22256868 Review. - Class-imbalanced classifiers for high-dimensional data.
Lin WJ, Chen JJ. Lin WJ, et al. Brief Bioinform. 2013 Jan;14(1):13-26. doi: 10.1093/bib/bbs006. Epub 2012 Mar 9. Brief Bioinform. 2013. PMID: 22408190 Review.
Cited by
- Quantitative Structure-Mutation-Activity Relationship Tests (QSMART) model for protein kinase inhibitor response prediction.
Huang LC, Yeung W, Wang Y, Cheng H, Venkat A, Li S, Ma P, Rasheed K, Kannan N. Huang LC, et al. BMC Bioinformatics. 2020 Nov 12;21(1):520. doi: 10.1186/s12859-020-03842-6. BMC Bioinformatics. 2020. PMID: 33183223 Free PMC article. - Improving drug response prediction by integrating multiple data sources: matrix factorization, kernel and network-based approaches.
Güvenç Paltun B, Mamitsuka H, Kaski S. Güvenç Paltun B, et al. Brief Bioinform. 2021 Jan 18;22(1):346-359. doi: 10.1093/bib/bbz153. Brief Bioinform. 2021. PMID: 31838491 Free PMC article. - Artificial Intelligence, Machine Learning, and Deep Learning in Real-Life Drug Design Cases.
Muller C, Rabal O, Diaz Gonzalez C. Muller C, et al. Methods Mol Biol. 2022;2390:383-407. doi: 10.1007/978-1-0716-1787-8_16. Methods Mol Biol. 2022. PMID: 34731478 Review. - Integration of Computational Docking into Anti-Cancer Drug Response Prediction Models.
Narykov O, Zhu Y, Brettin T, Evrard YA, Partin A, Shukla M, Xia F, Clyde A, Vasanthakumari P, Doroshow JH, Stevens RL. Narykov O, et al. Cancers (Basel). 2023 Dec 21;16(1):50. doi: 10.3390/cancers16010050. Cancers (Basel). 2023. PMID: 38201477 Free PMC article. - Simultaneous regression and classification for drug sensitivity prediction using an advanced random forest method.
Lenhof K, Eckhart L, Gerstner N, Kehl T, Lenhof HP. Lenhof K, et al. Sci Rep. 2022 Aug 5;12(1):13458. doi: 10.1038/s41598-022-17609-x. Sci Rep. 2022. PMID: 35931707 Free PMC article.
References
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases