Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID - PubMed (original) (raw)
Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID
Linda Warfield et al. Mol Cell. 2017.
Abstract
Previous studies suggested that expression of most yeast mRNAs is dominated by either transcription factor TFIID or SAGA. We re-examined the role of TFIID by rapid depletion of S. cerevisiae TFIID subunits and measurement of changes in nascent transcription. We find that transcription of nearly all mRNAs is strongly dependent on TFIID function. Degron-dependent depletion of Taf1, Taf2, Taf7, Taf11, and Taf13 showed similar transcription decreases for genes in the Taf1-depleted, Taf1-enriched, TATA-containing, and TATA-less gene classes. The magnitude of TFIID dependence varies with growth conditions, although this variation is similar genome-wide. Many studies have suggested differences in gene-regulatory mechanisms between TATA and TATA-less genes, and these differences have been attributed in part to differential dependence on SAGA or TFIID. Our work indicates that TFIID participates in expression of nearly all yeast mRNAs and that differences in regulation between these two gene categories is due to other properties.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
Figure 1
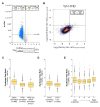
Auxin-dependent depletion of yeast TFIID subunits. (A) Listed at left are yeast TFIID subunits in lobe C, the shared or similar TFIID/SAGA core subunits and other auxiliary subunits. * indicates subunits tagged with the 3xV5-IAA7 degron tag and # indicates subunits tagged with FRB for anchor-away. Blue indicates subunits shared with SAGA and Orange indicates that SAGA contains a related but SAGA-specific subunit. Shown at right is Western analysis of cells treated with 3-IAA for the indicated time. Western blot was probed with anti V5 antibody. (B) Analysis of TFIID subunits after Taf depletion. After 30 min treatment with 3-IAA, whole cell extracts were made and immune precipitated with anti Taf4 antibody. Shown is quantitative Western analysis of the IP, probing for Tafs 1, 3, 4, 12 and TBP. * indicates a non-specific signal observed with the anti Taf1 antibody.
Figure 2
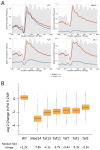
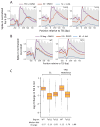
Genome-wide decreases in transcription upon depletion of TFIID and Mediator subunits. (A) Metagene profile of native Pol II ChIP in wild type and cells containing the indicated IAA7 degron fusion (4807 genes). Results are aligned at the major TSS (Park et al., 2014). Blue line = cells treated with 3-IAA for 30 min prior to harvest; red line = cells treated equivalently but with DMSO. Data are averages of biological duplicates with samples normalized by spike-in with S. pombe Rpb3-Flag cells. Grey shows metagene profile of histone H3 plotted using published H3 ChIP-seq data (Thurtle and Rine, 2014). The degron fused strains are labeled in the upper right corner of the different panels. (B) Box plots show the change in native Pol II ChIP on a log2 scale from 4807 genes comparing DMSO and 3-IAA addition. Median fold-changes in ChIP signals are indicated below. See also Fig S1 and Table S3.
Figure 3
TFIID functions at genes previously thought to be less dependent on TFIID. (A) Box plots of data from Fig 3 split into “Taf1-enriched” and “Taf1-depleted” categories (Rhee and Pugh, 2012). (B) Box plots of data from Fig 3 split into TATA-containing and TATA-less promoter categories (Rhee and Pugh, 2012).
Figure 4
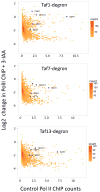
Several previously characterized Taf1-independent genes are dependent on other TFIID subunits. Plotted is a hexagonally binned 2d-histogram of Log2 fold change in Pol II ChIP upon 3-IAA addition vs. spike-normalized Pol II ChIP signal (a measure of transcription) averaged over the 1–100 bp from the TSS of each gene (4807 genes). The blue points are genes used in pre genome-wide studies to examine TFIID-dependence (Kuras et al., 2000; Li et al., 2000). See also Figs S1, S2 and Table S3.
Figure 5
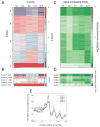
Analysis of genome-wide dependence on TFIID subunits. (A) Heatmap of genes ordered by clusters that were obtained after k-means clustering of the Z-scores from Taf-degron experiments. (B) The mean Z-scores of the 5 clusters for the 5 different Taf subunit degrons plotted as a heatmap. The mean values are indicated in each heatmap cell. (C) Same ordering of genes as (A), but the spike-normalized change in Pol II at the TSS is plotted. (D) The mean Spike-normalized change of the 5 clusters for the 5 different Taf subunit degrons plotted as a heatmap. (E) Average Taf1 ChEC-seq profiles (Grünberg et al., 2016) plotted for the 5 clusters shown in panel (A). See also Tables S3, S4, and S5.
Figure 6
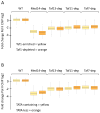
cDTA analysis of Taf4 anchor-away strain. (A) Volcano plot showing fold changes in newly-synthesized mRNA levels upon Taf4 nuclear depletion relative to their significance (_p_-value). 5657 genes were analyzed in two biological replicates and thresholds of 2-fold change and 0.05 _p_-values were considered (genes upregulated by 2-fold are indicated in yellow and genes downregulated by 2-fold are indicated in blue). (B) Changes in mRNA synthesis rates for 5434 genes upon rapamycin treatment in a TAF4-FRB strain were plotted against the changes of the decay rates. Log2 values of the average synthesis rate ratio and decay rate ratio are indicated. (C–E) Box plots showing the distribution of changes in synthesis rates upon Taf4 nuclear depletion according to the gene class (C, D) or to the gene expression levels (E) Dotted horizontal lines in panels C, D, E show the median of the synthesis rate ratio. See also Figs S3 and S4.
Figure 7
TFIID-dependence of transcription in synthetic media and heat shock stress. (A) Metagene profile of native Pol II ChIP from cells grown in synthetic complete (SC) media and containing the indicated IAA7 degron fusion. Results are presented as in Fig 2. Purple line is Pol II ChIP data from YPD-grown cells (+DMSO) from Fig 2; Red and blue lines are cells grown in SC with or without 3-IAA for 30 min. (B) Same as in A but Pol II ChIP from cells under heat shock stress. Purple line is Pol II ChIP from YPD-grown cells (+DMSO) in Fig 2. Red and blue lines are from YPD-grown cells heat shocked for 30 min prior to 30 min addition of DMSO or 3-IAA as indicated. (C) Genome-wide changes in transcription upon TFIID subunit depletion from the data in panels A and B. Box plots show the change in native Pol II ChIP on a log2 scale from 4807 genes comparing DMSO and 3-IAA addition. For heat shock conditions, the plot indicates changes in Pol II ChIP in heat shocked WT and heat shocked degron-containing cells upon addition of 3-IAA. Median fold-changes in ChIP signals are indicated below. See also Figs S5, S6 and S7.
Similar articles
- SAGA Is a General Cofactor for RNA Polymerase II Transcription.
Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, Tora L. Baptista T, et al. Mol Cell. 2017 Oct 5;68(1):130-143.e5. doi: 10.1016/j.molcel.2017.08.016. Epub 2017 Sep 14. Mol Cell. 2017. PMID: 28918903 Free PMC article. - Mechanistic Differences in Transcription Initiation at TATA-Less and TATA-Containing Promoters.
Donczew R, Hahn S. Donczew R, et al. Mol Cell Biol. 2017 Dec 13;38(1):e00448-17. doi: 10.1128/MCB.00448-17. Print 2018 Jan 1. Mol Cell Biol. 2017. PMID: 29038161 Free PMC article. - SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae.
Watanabe K, Kokubo T. Watanabe K, et al. PLoS One. 2017 Nov 27;12(11):e0188435. doi: 10.1371/journal.pone.0188435. eCollection 2017. PLoS One. 2017. PMID: 29176831 Free PMC article. - SAGA and TFIID: Friends of TBP drifting apart.
Timmers HTM. Timmers HTM. Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194604. doi: 10.1016/j.bbagrm.2020.194604. Epub 2020 Jul 14. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32673655 Review. - Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.
Pereira LA, Klejman MP, Timmers HT. Pereira LA, et al. Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
Cited by
- Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator.
Sanborn AL, Yeh BT, Feigerle JT, Hao CV, Townshend RJ, Lieberman Aiden E, Dror RO, Kornberg RD. Sanborn AL, et al. Elife. 2021 Apr 27;10:e68068. doi: 10.7554/eLife.68068. Elife. 2021. PMID: 33904398 Free PMC article. - TOR complex 2 contributes to regulation of gene expression via inhibiting Gcn5 recruitment to subtelomeric and DNA replication stress genes.
Cohen A, Pataki E, Kupiec M, Weisman R. Cohen A, et al. PLoS Genet. 2022 Feb 14;18(2):e1010061. doi: 10.1371/journal.pgen.1010061. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35157728 Free PMC article. - Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Antonova SV, Haffke M, Corradini E, Mikuciunas M, Low TY, Signor L, van Es RM, Gupta K, Scheer E, Vos HR, Tora L, Heck AJR, Timmers HTM, Berger I. Antonova SV, et al. Nat Struct Mol Biol. 2018 Dec;25(12):1119-1127. doi: 10.1038/s41594-018-0156-z. Epub 2018 Dec 3. Nat Struct Mol Biol. 2018. PMID: 30510221 Free PMC article. - The SAGA core module is critical during Drosophila oogenesis and is broadly recruited to promoters.
Soffers JHM, Alcantara SG, Li X, Shao W, Seidel CW, Li H, Zeitlinger J, Abmayr SM, Workman JL. Soffers JHM, et al. PLoS Genet. 2021 Nov 22;17(11):e1009668. doi: 10.1371/journal.pgen.1009668. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34807910 Free PMC article. - Disordered C-terminal domain drives spatiotemporal confinement of RNAPII to enhance search for chromatin targets.
Ling YH, Ye Z, Liang C, Yu C, Park G, Corden JL, Wu C. Ling YH, et al. bioRxiv [Preprint]. 2023 Nov 30:2023.07.31.551302. doi: 10.1101/2023.07.31.551302. bioRxiv. 2023. PMID: 37577667 Free PMC article. Updated. Preprint.
References
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
- Berger I, Blanco AG, Boelens R, Cavarelli J, Coll M, Folkers GE, Nie Y, Pogenberg V, Schultz P, Wilmanns M, et al. Structural insights into transcription complexes. Journal of Structural Biology. 2011;175:135–146. - PubMed
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases