Variations in oral microbiota associated with oral cancer - PubMed (original) (raw)
Clinical Trial
Variations in oral microbiota associated with oral cancer
Hongsen Zhao et al. Sci Rep. 2017.
Abstract
Individual bacteria and shifts in microbiome composition are associated with human disease, including cancer. To unravel the connections underlying oral bacterial dysbiosis and oral squamous cell carcinoma (OSCC), cancer lesion samples and anatomically matched normal samples were obtained from the same patients. We then profiled the bacteria within OSCC lesion surface samples at the species level using next-generation sequencing to comprehensively investigate bacterial community composition and functional genes in these samples. Significantly greater bacterial diversity was observed in the cancer samples than in the normal samples. Compared with previous studies, we identified many more taxa demonstrating remarkably different distributions between the groups. In particular, a group of periodontitis-correlated taxa, including Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella and Parvimonas, was significantly enriched in OSCC samples. Additionally, several operational taxonomic units (OTUs) associated with Fusobacterium were highly involved in OSCC and demonstrated good diagnostic power. Our study revealed drastic changes in surface bacterial communities of OSCC. The findings enrich knowledge of the association between oral bacterial communities and oral cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
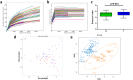
Figure 1
Comparison of oral microbiota structures in the N and T groups. (a) Rarefaction analysis of bacterial 16 S rRNA gene sequences was performed to evaluate whether further sequencing would likely detect additional taxa, indicated by a plateau. Different colors represent different samples. (b) Shannon index curves were constructed to evaluate the numbers of samples likely required to identify additional taxa, indicated by a plateau. Different colors represent different samples. (c) Box plots depict differences in bacterial diversity between the N and T groups according to the Shannon index. (d) PCA at the OTU level. (e) Partial least square discriminant score plot of oral microbiota between the N and T groups. N, clinical normal samples; T, oral cancer samples.
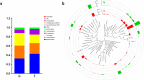
Figure 2
Composition of bacterial communities across samples at the phylum and genus levels. (a) Relative abundance of bacterial phyla among the N and T groups. (b) Classification tree of the 50 most abundant genera across all samples. The outer cycle colored bars represent the relative abundances of taxa in each group. N, clinical normal samples; T, oral cancer samples.
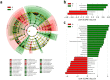
Figure 3
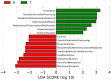
Distinct taxa identified in the N and T groups using LEfSe analysis. (a) Cladogram constructed using the LEfSe method to indicate the phylogenetic distribution of bacteria that were remarkably enriched in the N and T groups. (b) LDA scores showed significant bacterial differences within groups at the phylum level.(c) LDA scores showed significant bacterial differences within groups at the genus level. N, clinical normal samples; T, oral cancer samples.
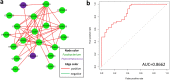
Figure 4
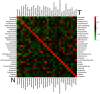
Co-occurrence network of Fusobacterium comprising OTUs and the diagnostic power of selected OTUs. (a) Each node represents an OTU colored for its genus-level phylotypes, and each edge represents a significant co-occurrence relationship colored according to its association (red: positive, green: negative). (b) ROC curves for selected Fusobacterium OTUs were constructed to predict diagnostic power.
Figure 5
Co-occurrence and co-exclusion analysis of bacterial genera. Pearson correlations among the top 30 most abundant bacterial genera were calculated and analyzed; groups are shown on the left and right. Correlation values ranged from −1.00 (green) to 1.00 (red).
Figure 6
LDA scores predict gene function enriched in different groups using PICRUSt. N, clinical normal samples; T, oral cancer samples.
Similar articles
- 16S rRNA based metagenomic analysis unveils unique oral microbial signatures in oral squamous cell carcinoma cases from Coastal Karnataka, India.
Aparna KG, Ravindra J, Chakraborty G, Ballamoole KK, Vinaya Kumar JR, Chakraborty A. Aparna KG, et al. Acta Microbiol Immunol Hung. 2024 Jul 1;71(3):253-262. doi: 10.1556/030.2024.02307. Print 2024 Sep 18. Acta Microbiol Immunol Hung. 2024. PMID: 38949882 - The Oral Microbiota May Have Influence on Oral Cancer.
Zhang L, Liu Y, Zheng HJ, Zhang CP. Zhang L, et al. Front Cell Infect Microbiol. 2020 Jan 15;9:476. doi: 10.3389/fcimb.2019.00476. eCollection 2019. Front Cell Infect Microbiol. 2020. PMID: 32010645 Free PMC article. - Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer.
Yang SF, Huang HD, Fan WL, Jong YJ, Chen MK, Huang CN, Chuang CY, Kuo YL, Chung WH, Su SC. Yang SF, et al. Oral Oncol. 2018 Feb;77:1-8. doi: 10.1016/j.oraloncology.2017.12.005. Epub 2017 Dec 9. Oral Oncol. 2018. PMID: 29362114 - The Role of the Microbiome in Oral Squamous Cell Carcinoma with Insight into the Microbiome-Treatment Axis.
Sami A, Elimairi I, Stanton C, Ross RP, Ryan CA. Sami A, et al. Int J Mol Sci. 2020 Oct 29;21(21):8061. doi: 10.3390/ijms21218061. Int J Mol Sci. 2020. PMID: 33137960 Free PMC article. Review. - Anaerobic bacterial communities associated with oral carcinoma: Intratumoral, surface-biofilm and salivary microbiota.
Minarovits J. Minarovits J. Anaerobe. 2021 Apr;68:102300. doi: 10.1016/j.anaerobe.2020.102300. Epub 2020 Nov 24. Anaerobe. 2021. PMID: 33246097 Review.
Cited by
- Microbiome analysis of saliva from oral squamous cell carcinoma (OSCC) patients and tobacco abusers with potential biomarkers for oral cancer screening.
Oyeyemi BF, Kaur US, Paramraj A, Chintamani, Tandon R, Kumar A, Bhavesh NS. Oyeyemi BF, et al. Heliyon. 2023 Nov 8;9(11):e21773. doi: 10.1016/j.heliyon.2023.e21773. eCollection 2023 Nov. Heliyon. 2023. PMID: 38034672 Free PMC article. - Looking beyond the smokescreen: can the oral microbiome be a tool or target in the management of tobacco-associated oral cancer?
Kadam S, Vandana M, Patwardhan S, Kaushik KS. Kadam S, et al. Ecancermedicalscience. 2021 Feb 1;15:1179. doi: 10.3332/ecancer.2021.1179. eCollection 2021. Ecancermedicalscience. 2021. PMID: 33777172 Free PMC article. Review. - Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review. - Microbial Changes Associated With Oral Cavity Cancer Progression.
Yan K, Auger S, Diaz A, Naman J, Vemulapalli R, Hasina R, Izumchenko E, Shogan B, Agrawal N. Yan K, et al. Otolaryngol Head Neck Surg. 2023 Jun;168(6):1443-1452. doi: 10.1002/ohn.211. Epub 2023 Jan 24. Otolaryngol Head Neck Surg. 2023. PMID: 36939272 Free PMC article. - Assessing the Effect of Smokeless Tobacco Consumption on Oral Microbiome in Healthy and Oral Cancer Patients.
Saxena R, Prasoodanan P K V, Gupta SV, Gupta S, Waiker P, Samaiya A, Sharma AK, Sharma VK. Saxena R, et al. Front Cell Infect Microbiol. 2022 Mar 31;12:841465. doi: 10.3389/fcimb.2022.841465. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35433507 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical