Development and optimization of a sensitive pseudovirus-based assay for HIV-1 neutralizing antibodies detection using A3R5 cells - PubMed (original) (raw)
Development and optimization of a sensitive pseudovirus-based assay for HIV-1 neutralizing antibodies detection using A3R5 cells
Qingqing Chen et al. Hum Vaccin Immunother. 2018.
Abstract
Sensitive assays for HIV-1 neutralizing antibody detection are urgently needed for vaccine immunogen optimization and identification of protective immune response levels. In this study, we developed an easy-to-use HIV-1 pseudovirus neutralization assay based on a human CD4+ lymphoblastoid cell line A3R5 by employing a high efficient pseudovirus production system. Optimal conditions for cell counts, infection time, virus dose and concentration of DEAE-dextran were tested and identified. For T-cell line-adapted tier 1 virus strains, significantly higher inhibitory efficiency was observed for both monoclonal neutralizing antibody (4 fold) and plasma (2 fold) samples in A3R5 than those in TZM-bl assay. For circulating tier 2 strains, the A3R5 pseudovirus assay showed even much higher sensitivity for both neutralizing antibody (10 fold) and plasma (9 fold) samples. When sequential neutralizing antibody seroconverting samples were tested in both A3R5 and TZM-bl assays, the seroconverting points could be detected earlier for tier 1 (15.7 weeks) and tier 2 (68.3 weeks) strains in A3R5 assay respectively. The high sensitive pseudovirus assay using more physiological target cells could serve as an alternative to the TZM-bl assay for evaluation of vaccine-induced neutralizing antibodies and identification of the correlates of protection.
Keywords: A3R5; HIV-1; TZM-bl; neutralizing antibody; pseudovirus.
Figures
Figure 1.
Optimization of cell numbers for the A3R5 pseudovirus assay. (A) Influence of cell numbers on neutralization results when tested against monoclonal neutralizing antibody samples (PG9 and 2F5) with pseudovirus 11036. (B) Influence of cell numbers on neutralization results when tested against serum samples (HB118 and BJ170) with 11036. (C) Effect of the cell density on the fitness of the neutralization curve.
Figure 2.
Optimization of the pseudovirus input for the A3R5 pseudovirus assay. (A) The neutralization curve under different pseudovirus inoculum. (B) Effect of the pseudovirus input amount on the fitness of the neutralization curve.
Figure 3.
Optimization of DEAE-dextran concentration for the A3R5 pseudovirus assay. Effect of the DEAE-dextran concentration on infectivity of the pseudovirus (A) and the viability of the A3R5 cells (B).
Figure 4.
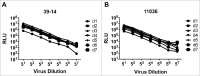
Determination of incubation time for the pseudovirus A3R5 assay. Relationship between the RLU and the incubation time for pseudovirus 39–14 (A) and 11306 (B).
Figure 5.
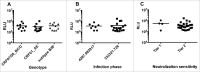
Infectivity of pseudoviruses to A3R5 cells. Pseudoviruses were diluted at 5 fold in triplicate and infected A3R5 cell. The average relative luminescent unit (RLU) values are indicated. (A) The infectivity of pseudoviruses from different subtypes. (B) The infectivity of pseudoviruses from different infection phases. (C) The infectivity of pseudoviruses with different neutralization sensitivities.
Figure 6.
Validation of the pseudovirus A3R5 assay. (A) specificity: 20 HIV-1-negative plasma samples were tested against a panel of seven pseudoviruses (SF162, Bal26 from tier 1 subtype B; the remaining from tier 2 strains, 11317 from subtype C; 11056, 11058 from subtype B; BJ5.11, GX24.8 from CRF01_AE). (B) Reproducibility: two plasma samples (HJ182 and HB188) were employed to test against pseudovirus 11036. Each plasma sample was tested ten times in three independent runs by different operators.
Figure 7.
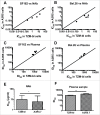
Comparison of the pseudovirus A3R5 assay with the TZM-bl assay tested against tier 1 pseudoviruses. Five NAbs (2F5, 4E10, PG16, 2G12 and b12) were tested against two pseudoviruses SF162 (A) and Bal.26 (B). Five plasma samples (HB4, HB118, HB120, BJ182 and TJ208) were tested against the same two pseudoviruses SF162 (C) and Bal.26 (D). The diagonal line depicts x = y (TZM-bl ID50/IC50 = A3R5 ID50/IC50 values). Statistical analyses for NAb (E) and plasma (F) samples were conducted respectively (Wilcoxon matched pairs test, * for p<0.05, and ** for p < 0.01).
Figure 8.
Comparison of the pseudovirus A3R5 assay with the TZM-bl assay tested against tier 2 pseudoviruses. Five NAbs were tested against four tier 2 pseudoviruses 11036 (A), 11058 (B) and 11317 (C), and 11506 (D). Five plasma samples were tested against the same four pseudoviruses 11036 (E), 11058 (F) and 11317 (G), and 11506 (H). The diagonal line depicts x = y (TZM-bl ID50/IC50 = A3R5 ID50/IC50 values). Statistical analyses for NAb (I) and plasma (J) samples were conducted respectively (Wilcoxon matched pairs test, *** for p<0.001=.
Figure 9.
Comparison of A3R5 and TZM-bl assay using NAb seroconverting samples against tier 1 pseudoviurs. Serial NAb seroconverting plasma samples collected from 8 CRF01_AE-infected individuals were tested against pseudovirus SF162 in both A3R5 and TZM-bl assays. The dash and full lines indicate the cutoff values for A3R5 and TZM-bl assay respectively.
Figure 10.
Comparison of A3R5 and TZM-bl assay using NAb seroconverting samples against tier 2 pseudoviurses. Serial NAb seroconverting plasma samples were tested against 3 tier 2 pseudoviruses in both A3R5 and TZM-bl assays. The dash and full lines indicate the cutoff values for A3R5 and TZM-bl assay respectively.
Similar articles
- Optimization and validation of a neutralizing antibody assay for HIV-1 in A3R5 cells.
Sarzotti-Kelsoe M, Daniell X, Todd CA, Bilska M, Martelli A, LaBranche C, Perez LG, Ochsenbauer C, Kappes JC, Rountree W, Denny TN, Montefiori DC. Sarzotti-Kelsoe M, et al. J Immunol Methods. 2014 Jul;409:147-60. doi: 10.1016/j.jim.2014.02.013. Epub 2014 Mar 6. J Immunol Methods. 2014. PMID: 24607608 Free PMC article. - Optimization and proficiency testing of a pseudovirus-based assay for detection of HIV-1 neutralizing antibody in China.
Nie J, Wang W, Wen Z, Song A, Hong K, Lu S, Zhong P, Xu J, Kong W, Li J, Shang H, Ling H, Ruan L, Wang Y. Nie J, et al. J Virol Methods. 2012 Nov;185(2):267-75. doi: 10.1016/j.jviromet.2012.07.011. Epub 2012 Jul 13. J Virol Methods. 2012. PMID: 22796285 - Inhibitory Effect of Individual or Combinations of Broadly Neutralizing Antibodies and Antiviral Reagents against Cell-Free and Cell-to-Cell HIV-1 Transmission.
Gombos RB, Kolodkin-Gal D, Eslamizar L, Owuor JO, Mazzola E, Gonzalez AM, Korioth-Schmitz B, Gelman RS, Montefiori DC, Haynes BF, Schmitz JE. Gombos RB, et al. J Virol. 2015 Aug;89(15):7813-28. doi: 10.1128/JVI.00783-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995259 Free PMC article. - Impact of host cell variation on the neutralization of HIV-1 in vitro.
Polonis VR, Schuitemaker H, Bunnik EM, Brown BK, Scarlatti G. Polonis VR, et al. Curr Opin HIV AIDS. 2009 Sep;4(5):400-7. doi: 10.1097/COH.0b013e32832edc50. Curr Opin HIV AIDS. 2009. PMID: 20048704 Review. - Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination.
Polonis VR, Brown BK, Rosa Borges A, Zolla-Pazner S, Dimitrov DS, Zhang MY, Barnett SW, Ruprecht RM, Scarlatti G, Fenyö EM, Montefiori DC, McCutchan FE, Michael NL. Polonis VR, et al. Virology. 2008 Jun 5;375(2):315-20. doi: 10.1016/j.virol.2008.02.007. Epub 2008 Mar 25. Virology. 2008. PMID: 18367229 Review.
Cited by
- Nipah pseudovirus system enables evaluation of vaccines in vitro and in vivo using non-BSL-4 facilities.
Nie J, Liu L, Wang Q, Chen R, Ning T, Liu Q, Huang W, Wang Y. Nie J, et al. Emerg Microbes Infect. 2019;8(1):272-281. doi: 10.1080/22221751.2019.1571871. Emerg Microbes Infect. 2019. PMID: 30866781 Free PMC article. - Characterization of SARS-CoV-2 worldwide transmission based on evolutionary dynamics and specific viral mutations in the spike protein.
Liu J, Chen X, Liu Y, Lin J, Shen J, Zhang H, Yin J, Pu R, Ding Y, Cao G. Liu J, et al. Infect Dis Poverty. 2021 Aug 21;10(1):112. doi: 10.1186/s40249-021-00895-4. Infect Dis Poverty. 2021. PMID: 34419160 Free PMC article. - Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro.
Yang R, Huang B, A R, Li W, Wang W, Deng Y, Tan W. Yang R, et al. Biosaf Health. 2020 Dec;2(4):226-231. doi: 10.1016/j.bsheal.2020.08.004. Epub 2020 Aug 21. Biosaf Health. 2020. PMID: 32864605 Free PMC article. - SARS-CoV-2-specific CD4+ T cells are associated with long-term persistence of neutralizing antibodies.
Wang Z, Yang X, Mei X, Zhou Y, Tang Z, Li G, Zhong J, Yu M, Huang M, Su X, Lin B, Cao P, Yang J, Ran P. Wang Z, et al. Signal Transduct Target Ther. 2022 Apr 23;7(1):132. doi: 10.1038/s41392-022-00978-0. Signal Transduct Target Ther. 2022. PMID: 35461307 Free PMC article. - Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals.
Zhang J, Wu Q, Liu Z, Wang Q, Wu J, Hu Y, Bai T, Xie T, Huang M, Wu T, Peng D, Huang W, Jin K, Niu L, Guo W, Luo D, Lei D, Wu Z, Li G, Huang R, Lin Y, Xie X, He S, Deng Y, Liu J, Li W, Lu Z, Chen H, Zeng T, Luo Q, Li YP, Wang Y, Liu W, Qu X. Zhang J, et al. Nat Microbiol. 2021 Jan;6(1):51-58. doi: 10.1038/s41564-020-00824-5. Epub 2020 Nov 16. Nat Microbiol. 2021. PMID: 33199863
References
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al.. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285-9. doi:10.1126/science.1178746. PMID:19729618 - DOI - PMC - PubMed
- Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15(8):866-70. PMID:19525964 - PubMed
- Ferrantelli F, Rasmussen RA, Buckley KA, Li PL, Wang T, Montefiori DC, Katinger H, Stiegler G, Anderson DC, McClure HM, et al.. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian-human immunodeficiency virus by human anti-HIV monoclonal antibodies. J Infect Dis. 2004;189(12):2167-73. doi:10.1086/420833. PMID:15181562 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials