Investigating Difficult Nodes in the Placental Mammal Tree with Expanded Taxon Sampling and Thousands of Ultraconserved Elements - PubMed (original) (raw)
Investigating Difficult Nodes in the Placental Mammal Tree with Expanded Taxon Sampling and Thousands of Ultraconserved Elements
Jacob A Esselstyn et al. Genome Biol Evol. 2017.
Abstract
The phylogeny of eutherian mammals contains some of the most recalcitrant nodes in the tetrapod tree of life. We combined comprehensive taxon and character sampling to explore three of the most debated interordinal relationships among placental mammals. We performed in silico extraction of ultraconserved element loci from 72 published genomes and invitro enrichment and sequencing of ultraconserved elements from 28 additional mammals, resulting in alignments of 3,787 loci. We analyzed these data using concatenated and multispecies coalescent phylogenetic approaches, topological tests, and exploration of support among individual loci to identify the root of Eutheria and the sister groups of tree shrews (Scandentia) and horses (Perissodactyla). Individual loci provided weak, but often consistent support for topological hypotheses. Although many gene trees lacked accepted species-tree relationships, summary coalescent topologies were largely consistent with inferences from concatenation. At the root of Eutheria, we identified consistent support for a sister relationship between Xenarthra and Afrotheria (i.e., Atlantogenata). At the other nodes of interest, support was less consistent. We suggest Scandentia is the sister of Primatomorpha (Euarchonta), but we failed to reject a sister relationship between Scandentia and Glires. Similarly, we suggest Perissodactyla is sister to Cetartiodactyla (Euungulata), but a sister relationship between Perissodactyla and Chiroptera remains plausible.
Keywords: Atlantogenata; Eutheria; Perissodactyla; Scandentia; phylogenomics.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
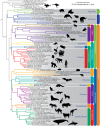
Fig. 1.
—Maximum likelihood estimate of mammalian phylogeny from 3,787 concatenated ultraconserved elements. Clades discussed in the text are labeled. Branches between Metatheria (marsupials), Eutheria (placentals), and Prototheria (platypus; not shown) were truncated for ease of presentation. Bootstrap support values are 100 unless otherwise noted. Taxa with an asterisk after the specific epithet are those we enriched and sequenced.
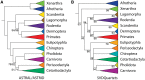
Fig. 2.
—Species tree estimates of eutherian relationships derived from analysis of 3,787 ultraconserved elements in (A) ASTRAL and ASTRID, and (B) SVDquartets. The topology is collapsed to the ordinal level or higher. Bootstrap support values are 100 unless otherwise noted.
Fig. 3.
—Summary of the consistency of individual gene trees with competing topological hypotheses for species relationships. (A) Hypotheses regarding earliest divergence among placentals, (B) relationships of Scandentia, and (C) relationships of Perissodactyla. The percentage of gene trees consistent with a multi-constraint topological filter is shown above each topology (e.g., 24.7% of gene trees contain monophyletic Afrotheria, Xenarthra, and Afrotheria + Xenarthra). The percentage of gene trees consistent with a single-constraint topological filter is shown to the right of the relevant node (e.g., 26.3% contain a monophyletic Afrotheria + Xenarthra). Average bootstrap support (BS) at a particular bifurcation are shown to the left of each node (e.g., 33.8% mean BS for Afrotheria + Xenarthra). In each panel, relevant tip labels include the percent of gene trees containing that clade, followed by their average BS at that node among those gene trees (e.g., Afrotheria found in 73% of gene trees with mean of 59% BS).
Fig. 4.
—Plots of per gene ΔlnL against the number of parsimony informative sites for our maximum likelihood (ML) tree (Hypothesis 1) versus 2–4 alternative hypotheses (Hypotheses 2–5). Each topological hypothesis is defined in the gray box at center right. The following orders are abbreviated: Perissodactyla (Per); Cetartiodactyla (Cet); Chiroptera (Chir); and Carnivora (Carn).
Similar articles
- Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis.
McCormack JE, Faircloth BC, Crawford NG, Gowaty PA, Brumfield RT, Glenn TC. McCormack JE, et al. Genome Res. 2012 Apr;22(4):746-54. doi: 10.1101/gr.125864.111. Epub 2011 Dec 29. Genome Res. 2012. PMID: 22207614 Free PMC article. - Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model.
Song S, Liu L, Edwards SV, Wu S. Song S, et al. Proc Natl Acad Sci U S A. 2012 Sep 11;109(37):14942-7. doi: 10.1073/pnas.1211733109. Epub 2012 Aug 28. Proc Natl Acad Sci U S A. 2012. PMID: 22930817 Free PMC article. - The gene tree delusion.
Springer MS, Gatesy J. Springer MS, et al. Mol Phylogenet Evol. 2016 Jan;94(Pt A):1-33. doi: 10.1016/j.ympev.2015.07.018. Epub 2015 Jul 31. Mol Phylogenet Evol. 2016. PMID: 26238460 - Mammal madness: is the mammal tree of life not yet resolved?
Foley NM, Springer MS, Teeling EC. Foley NM, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20150140. doi: 10.1098/rstb.2015.0140. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325836 Free PMC article. Review. - The chromosomes of Afrotheria and their bearing on mammalian genome evolution.
Svartman M, Stanyon R. Svartman M, et al. Cytogenet Genome Res. 2012;137(2-4):144-53. doi: 10.1159/000341387. Epub 2012 Aug 3. Cytogenet Genome Res. 2012. PMID: 22868637 Review.
Cited by
- Accurate, scalable, and fully automated inference of species trees from raw genome assemblies using ROADIES.
Gupta A, Mirarab S, Turakhia Y. Gupta A, et al. bioRxiv [Preprint]. 2024 Jun 1:2024.05.27.596098. doi: 10.1101/2024.05.27.596098. bioRxiv. 2024. PMID: 38854139 Free PMC article. Preprint. - Basicranial evidence suggests picrodontid mammals are not stem primates.
Crowell JW, Wible JR, Chester SGB. Crowell JW, et al. Biol Lett. 2024 Jan;20(1):20230335. doi: 10.1098/rsbl.2023.0335. Epub 2024 Jan 10. Biol Lett. 2024. PMID: 38195058 - Improved mammalian family phylogeny using gap-rare multiple sequence alignment: A timetree of extant placentals and marsupials.
Liu GM, Pan Q, Du J, Zhu PF, Liu WQ, Li ZH, Wang L, Hu CY, Dai YC, Zhang XX, Zhang Z, Yu Y, Li M, Wang PC, Wang X, Li M, Zhou XM. Liu GM, et al. Zool Res. 2023 Nov 18;44(6):1064-1079. doi: 10.24272/j.issn.2095-8137.2023.189. Zool Res. 2023. PMID: 37914522 Free PMC article. - The evolution of antimicrobial peptides in Chiroptera.
Castellanos FX, Moreno-Santillán D, Hughes GM, Paulat NS, Sipperly N, Brown AM, Martin KR, Poterewicz GM, Lim MCW, Russell AL, Moore MS, Johnson MG, Corthals AP, Ray DA, Dávalos LM. Castellanos FX, et al. Front Immunol. 2023 Sep 26;14:1250229. doi: 10.3389/fimmu.2023.1250229. eCollection 2023. Front Immunol. 2023. PMID: 37822944 Free PMC article. - An atlas of rabbit development as a model for single-cell comparative genomics.
Ton MN, Keitley D, Theeuwes B, Guibentif C, Ahnfelt-Rønne J, Andreassen TK, Calero-Nieto FJ, Imaz-Rosshandler I, Pijuan-Sala B, Nichols J, Benito-Gutiérrez È, Marioni JC, Göttgens B. Ton MN, et al. Nat Cell Biol. 2023 Jul;25(7):1061-1072. doi: 10.1038/s41556-023-01174-0. Epub 2023 Jun 15. Nat Cell Biol. 2023. PMID: 37322291
References
- Bininda-Emonds ORP, et al.2007. The delayed rise of present-day mammals. Nature 446:507–512. - PubMed
- Blom MPK, Bragg JG, Potter S, Moritz C.. 2017. Accounting for uncertainty in gene tree estimation: summary-coalescent species tree inference in a challenging radiation of Australian lizards. Syst Biol. 66:352–366. - PubMed
- Brown JM, Thomson RC.. 2016. Bayes factors unmask highly variable information content, bias, and extreme influence in phylogenomic analyses. Syst Biol. 66(4):517–530. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources